| FourU | |
|---|---|
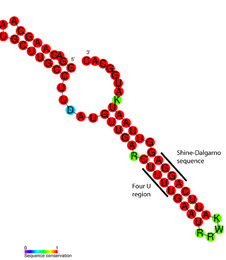 Consensus secondary structure of FourU RNA thermometers. Red colouring indicates the highest levels of nucleotide conservation. | |
| Identifiers | |
| Symbol | FourU |
| Rfam | RF01795 |
| Other data | |
| RNA type | RNA thermometer |
| Domain(s) | Salmonella; |
| PDB structures | PDBe |
FourU thermometers are a class of non-coding RNA thermometers found in Salmonella.[1] They are named 'FourU' due to the four highly conserved uridine nucleotides found directly opposite the Shine-Dalgarno sequence on hairpin II (pictured). RNA thermometers such as FourU control regulation of temperature via heat shock proteins in many prokaryotes.[2][3][4] FourU thermometers are relatively small RNA molecules, only 57 nucleotides in length, and have a simple two-hairpin structure.[1]
FourU are found in the 5' untranslated region of the gene for heat shock protein Salmonella agsA,[1][5][6] they repress translation of this protein by base-pairing the Shine-Dalgarno sequence of the gene's mRNA.[2] This prevents ribosomes from binding the start codon of the gene.[7]
They are also found in the 5'UTR of htrA (high temperature requirement) genes in Salmonella and E.coli.[8]
In V. cholerae fourU thermometer in the 5' of toxT controls its temperature-dependent translation. At human body temperature, the thermometer structure opens and to allow transcriptional activator protein ToxT translation, facilitating V. cholerae virulence.[9]
Other known RNA thermometers include the ROSE element[10][11] and Hsp90 cis-reg element.[12]
Response to temperature[edit]
Hairpin II appears to be a dynamic feature of FourU's secondary structure.[1][2] It undergoes a conformational shift when exposed to temperatures above 45 °C, becoming increasingly unpaired as temperature rises.[1] Hairpin I, in contrast, remains stably base-paired in temperatures as high as 50 °C, which implies the structural shift of hairpin II from closed to open may have an important role in heat shock response.[1] A later study used mutant analysis and calculations of enthalpy and entropy to support a cooperative zipper-type unfolding mechanism of FourU hairpin II in response to temperature increase.[2]
Sigma factor cooperation[edit]
Like other RNA thermometers, FourU is not solely responsible for temperature-dependent expression of its adjacent gene.[13] Instead, it operates in conjunction with a sigma factor (σ32)[14] which is known to also regulate many other genes.[15] Sigma factor-RNA thermometer combinations have been found to regulate other heat-shock genes (such as ibpA in E. coli)[4] which has led to speculation[by whom?] of undiscovered RNA thermometers operating alongside sigma factor modules to regulate other related genes as an additional level of control. Further speculation suggests the simpler RNA thermometer method of gene regulation may have evolved prior to the more complex sigma factor transcription control.[1]
agsA function[edit]
The agsA gene, which is regulated by FourU thermometers, was first discovered in Salmonella enterica.[6] The protein coded for by this gene is a small heat shock protein (sHSP) which protects bacteria from irreversible aggregation of proteins and aids in their refolding.[14] Mutant analysis confirmed the importance of agsA: a plasmid containing the gene and a promoter increased the survival rate of a thermosenstive mutant phenotype by remedying protein aggregation at high temperatures.[6] It has a similar function to the human chaperone α-crystallin.[16]
See also[edit]
References[edit]
- ^ a b c d e f g Waldminghaus T, Heidrich N, Brantl S, Narberhaus F (July 2007). "FourU: a novel type of RNA thermometer in Salmonella". Molecular Microbiology. 65 (2): 413–424. doi:10.1111/j.1365-2958.2007.05794.x. PMID 17630972.
- ^ a b c d Rinnenthal J, Klinkert B, Narberhaus F, Schwalbe H (June 2010). "Direct observation of the temperature-induced melting process of the Salmonella fourU RNA thermometer at base-pair resolution". Nucleic Acids Research. 38 (11): 3834–3847. doi:10.1093/nar/gkq124. PMC 2887971. PMID 20211842.
- ^ Narberhaus F, Waldminghaus T, Chowdhury S (January 2006). "RNA thermometers". FEMS Microbiology Reviews. 30 (1): 3–16. doi:10.1111/j.1574-6976.2005.004.x. PMID 16438677.
- ^ a b Waldminghaus T, Fippinger A, Alfsmann J, Narberhaus F (December 2005). "RNA thermometers are common in alpha- and gamma-proteobacteria". Biological Chemistry. 386 (12): 1279–1286. doi:10.1515/BC.2005.145. PMID 16336122. S2CID 84557068.
- ^ "aggregation suppressing protein". National Center for Biotechnology Information.
- ^ a b c Tomoyasu T, Takaya A, Sasaki T, Nagase T, Kikuno R, Morioka M, Yamamoto T (November 2003). "A new heat shock gene, AgsA, which encodes a small chaperone involved in suppressing protein aggregation in Salmonella enterica serovar typhimurium". Journal of Bacteriology. 185 (21): 6331–6339. doi:10.1128/JB.185.21.6331-6339.2003. PMC 219406. PMID 14563868.
- ^ Shine J, Dalgarno L (March 1975). "Determinant of cistron specificity in bacterial ribosomes". Nature. 254 (5495): 34–38. doi:10.1038/254034a0. PMID 803646. S2CID 4162567.
- ^ Klinkert B, Cimdins A, Gaubig LC, Roßmanith J, Aschke-Sonnenborn U, Narberhaus F (July 2012). "Thermogenetic tools to monitor temperature-dependent gene expression in bacteria". Journal of Biotechnology. 160 (1–2): 55–63. doi:10.1016/j.jbiotec.2012.01.007. PMID 22285954.
- ^ Weber GG, Kortmann J, Narberhaus F, Klose KE (September 2014). "RNA thermometer controls temperature-dependent virulence factor expression in Vibrio cholerae". Proceedings of the National Academy of Sciences of the United States of America. 111 (39): 14241–14246. doi:10.1073/pnas.1411570111. PMC 4191814. PMID 25228776.
- ^ Nocker A, Hausherr T, Balsiger S, Krstulovic NP, Hennecke H, Narberhaus F (December 2001). "A mRNA-based thermosensor controls expression of rhizobial heat shock genes". Nucleic Acids Research. 29 (23): 4800–4807. doi:10.1093/nar/29.23.4800. PMC 96696. PMID 11726689.
- ^ Waldminghaus T, Gaubig LC, Narberhaus F (November 2007). "Genome-wide bioinformatic prediction and experimental evaluation of potential RNA thermometers". Molecular Genetics and Genomics. 278 (5): 555–564. doi:10.1007/s00438-007-0272-7. PMID 17647020. S2CID 24747327.
- ^ Ahmed R, Duncan RF (November 2004). "Translational regulation of Hsp90 mRNA. AUG-proximal 5'-untranslated region elements essential for preferential heat shock translation". The Journal of Biological Chemistry. 279 (48): 49919–49930. doi:10.1074/jbc.M404681200. PMID 15347681.
- ^ Johansson J, Mandin P, Renzoni A, Chiaruttini C, Springer M, Cossart P (September 2002). "An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes". Cell. 110 (5): 551–561. doi:10.1016/S0092-8674(02)00905-4. PMID 12230973.
- ^ a b Bukau B (August 1993). "Regulation of the Escherichia coli heat-shock response". Molecular Microbiology. 9 (4): 671–680. doi:10.1111/j.1365-2958.1993.tb01727.x. PMID 7901731. S2CID 39147026.
- ^ Permina EA, Gelfand MS (2003). "Heat shock (sigma32 and HrcA/CIRCE) regulons in beta-, gamma- and epsilon-proteobacteria". Journal of Molecular Microbiology and Biotechnology. 6 (3–4): 174–181. doi:10.1159/000077248. PMID 15153770. S2CID 84915084.
- ^ Rajaraman K, Raman B, Ramakrishna T, Rao CM (May 2001). "Interaction of human recombinant alphaA- and alphaB-crystallins with early and late unfolding intermediates of citrate synthase on its thermal denaturation". FEBS Letters. 497 (2–3): 118–123. doi:10.1016/S0014-5793(01)02451-6. PMID 11377425.
Further reading[edit]
- Vogel J (January 2009). "A rough guide to the non-coding RNA world of Salmonella". Molecular Microbiology. 71 (1): 1–11. doi:10.1111/j.1365-2958.2008.06505.x. hdl:11858/00-001M-0000-000E-C124-A. PMID 19007416. S2CID 205366563.
- Jin H, Zhao Q, Gonzalez de Valdivia EI, Ardell DH, Stenström M, Isaksson LA (April 2006). "Influences on gene expression in vivo by a Shine-Dalgarno sequence". Molecular Microbiology. 60 (2): 480–492. doi:10.1111/j.1365-2958.2006.05110.x. PMID 16573696. S2CID 5686240.
- Chowdhury S, Ragaz C, Kreuger E, Narberhaus F (November 2003). "Temperature-controlled structural alterations of an RNA thermometer". The Journal of Biological Chemistry. 278 (48): 47915–47921. doi:10.1074/jbc.M306874200. PMID 12963744.
- Kaempfer R (November 2003). "RNA sensors: novel regulators of gene expression". EMBO Reports. 4 (11): 1043–1047. doi:10.1038/sj.embor.embor7400005. PMC 1326375. PMID 14593443.