WP:CHECKWIKI error fix #26. Convert HTML to wikicode. Do general fixes and cleanup if needed. - using AWB (12130) |
→Cladogram: alternative Tag: Visual edit |
||
| Line 117: | Line 117: | ||
|2=[[SAR supergroup|SAR]]}}}}}}}}}}}}}} |
|2=[[SAR supergroup|SAR]]}}}}}}}}}}}}}} |
||
}}}}|label1=[[Asgard Archaea]]}} |
}}}}|label1=[[Asgard Archaea]]}} |
||
In this view, excavata is highly paraphyletic, and is proposed to be abandoned.<ref>{{Cite journal|last=Cavalier-Smith|first=Thomas|date=2016-10-01|title=Higher classification and phylogeny of Euglenozoa|url=http://www.sciencedirect.com/science/article/pii/S0932473916300839|journal=European Journal of Protistology|volume=56|pages=250–276|doi=10.1016/j.ejop.2016.09.003}}</ref> |
In this view, excavata is highly paraphyletic, and is proposed to be abandoned.<ref>{{Cite journal|last=Cavalier-Smith|first=Thomas|date=2016-10-01|title=Higher classification and phylogeny of Euglenozoa|url=http://www.sciencedirect.com/science/article/pii/S0932473916300839|journal=European Journal of Protistology|volume=56|pages=250–276|doi=10.1016/j.ejop.2016.09.003}}</ref> In alternative view, the Discoba are sister to the rest of the Diphoda.<ref>{{Cite journal|last=Derelle|first=Romain|last2=Torruella|first2=Guifré|last3=Klimeš|first3=Vladimír|last4=Brinkmann|first4=Henner|last5=Kim|first5=Eunsoo|last6=Vlček|first6=Čestmír|last7=Lang|first7=B. Franz|last8=Eliáš|first8=Marek|date=2015-02-17|title=Bacterial proteins pinpoint a single eukaryotic root|url=http://www.pnas.org/content/112/7/E693|journal=Proceedings of the National Academy of Sciences|language=en|volume=112|issue=7|pages=E693–E699|doi=10.1073/pnas.1420657112|issn=0027-8424|pmc=PMC4343179|pmid=25646484}}</ref> |
||
== References == |
== References == |
||
Revision as of 17:56, 21 January 2017
| Excavata | |
|---|---|
| |
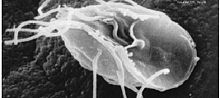
| Giardia lamblia, a parasitic diplomonad | |
| Scientific classification | |
| Domain: | Eukaryota |
| (unranked): | Excavata (Cavalier-Smith), 2002 |
| Phyla | |

Excavata is a major supergroup of unicellular organisms belonging to the domain Eukaryota.[1][2][3] Introduced by Thomas Cavalier-Smith in 2002 as a new phylogenetic category, it contains a variety of free-living and symbiotic forms, and also includes some important parasites of humans. Excavates were formerly considered to be included in the now obsolete Protista kingdom.[4] They are classified based on their flagellar structures,[5] and they are considered to be the oldest members (basal lineage) of flagellated organisms.[6]
The Discobaean excavata (Euglenozoa, Percolozoa, Tsukubea, Jakobea) now appear to be basal Eukaryotes, while the Metamonadaean and Malawimonaen excavata now appear to be sister clades of the Podiata. The Discoba may be highly paraphyletic.[7]
Characteristics
Many excavates lack 'classical' mitochondria—these organisms are often referred to as 'amitochondriate', although most retain a mitochondrial organelle in greatly modified form (e.g. a hydrogenosome or mitosome). Among those with mitochondria, the mitochondrial cristae may be tubular, discoidal, or in some cases, laminar. Most excavates have two, four, or more flagella[8] and many have a conspicuous ventral feeding groove with a characteristic ultrastructure, supported by microtubules. However, various groups that lack these traits may be considered excavates based on genetic evidence (primarily phylogenetic trees of molecular sequences).[4]
The closest that the excavates come to multicellularity are the Acrasidae slime molds. Like other cellular slime molds, they live most of their life as single cells, but will sometimes assemble into a larger cluster.
Classification
Excavates are classified into six major subdivisions at the phylum/class level. These are shown in the table below. An additional organism, Malawimonas, may also be included amongst excavates, though phylogenetic evidence is equivocal.
| Superphylum | Phylum/Class | Representative genera (examples) | Description |
|---|---|---|---|
| Discoba or JEH | Tsukubea | T. globosa | |
| Euglenozoa | Euglena, Trypanosoma | Many important parasites, one large group with plastids (chloroplasts) | |
| Heterolobosea (Percolozoa) | Naegleria, Acrasis | Most alternate between flagellate and amoeboid forms | |
| Jakobea | Jakoba, Reclinomonas | Free-living, sometimes loricate flagellates, with very gene-rich mitochondrial genomes | |
| Metamonada or POD | Preaxostyla | Oxymonads, Trimastix | Amitochondriate flagellates, either free-living (Trimastix) or living in the hindguts of insects |
| Fornicata | Giardia, Carpediemonas | Amitochondriate, mostly symbiotes and parasites of animals. | |
| Parabasalia | Trichomonas | Amitochondriate flagellates, generally intestinal commensals of insects. Some human pathogens. |
Discoba or JEH clade
Euglenozoa and Heterolobosea (Percolozoa) appear to be particularly close relatives, and are united by the presence of discoid cristae within the mitochondria (Superphylum Discicristata). More recently a close relationship has been shown between Discicristata and Jakobida,[9] the latter having tubular cristae like most other protists, and hence were united under the taxon name Discoba which was proposed for this apparently monophyletic group.[2]
Metamonads
Metamonads are unusual in having lost classical mitochondria—instead they have hydrogenosomes, mitosomes or uncharacterised organelles. The oxymonad Monocercomonoides is reported to have completely lost homologous organelles.
Monophyly
Excavate relationships are still uncertain; it is possible that they are not a monophyletic group. The monophyly of the excavates is far from clear, although it seems like there are several clades within the excavates which are monophyletic.[10]
Certain excavates are often considered among the most primitive eukaryotes, based partly on their placement in many evolutionary trees. This could encourage proposals that excavates are a paraphyletic grade that includes the ancestors of other living eukaryotes. However, the placement of certain excavates as 'early branches' may be an analysis artifact caused by long branch attraction, as has been seen with some other groups, for example, microsporidia.
Malawimonas
In addition to the groups mentioned in the table above, the genus Malawimonas is generally considered to be a member of Excavata owing to its typical excavate morphology, and phylogenetic affinity to excavate groups in some molecular phylogenies. However, its position among excavates remains elusive.[3]
Cladogram
A proposed cladogram for the positioning of the Excavata is, with the Eukaryote root in the excavates and the Eukaryotes as sister of the Heimdallarchaeota,[7][11][12][13][14][15][16][17]
In this view, excavata is highly paraphyletic, and is proposed to be abandoned.[18] In alternative view, the Discoba are sister to the rest of the Diphoda.[19]
References
- ^ Hampl, V.; Hug, L.; Leigh, J. W.; Dacks, J. B.; Lang, B. F.; Simpson, A. G. B.; Roger, A. J. (2009). "Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic "supergroups"". Proceedings of the National Academy of Sciences. 106 (10): 3859–3864. doi:10.1073/pnas.0807880106. PMC 2656170. PMID 19237557.
- ^ a b Hampl V, Hug L, Leigh JW, et al. (2009). "Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic "supergroups"". Proc. Natl. Acad. Sci. U.S.A. 106 (10): 3859–64. Bibcode:2009PNAS..106.3859H. doi:10.1073/pnas.0807880106. PMC 2656170. PMID 19237557.
- ^ a b Simpson, Ag; Inagaki, Y; Roger, Aj (2006). "Comprehensive multigene phylogenies of excavate protists reveal the evolutionary positions of "primitive" eukaryotes" (Free full text). Molecular Biology and Evolution. 23 (3): 615–25. doi:10.1093/molbev/msj068. PMID 16308337.
- ^ a b Cavalier-Smith, T (2002). "The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa". International Journal of Systematic and Evolutionary Microbiology. 52 (2): 297–354. doi:10.1099/00207713-52-2-297. PMID 11931142.
- ^ Simpson, AG (2003). "Cytoskeletal organization, phylogenetic affinities and systematics in the contentious taxon Excavata (Eukaryota)". International Journal of Systematic and Evolutionary Microbiology. 53 (Pt 6): 1759–1777. doi:10.1099/ijs.0.02578-0. PMID 14657103.
- ^ Dawson, Scott C; Paredez, Alexander R (2013). "Alternative cytoskeletal landscapes: cytoskeletal novelty and evolution in basal excavate protists". Current Opinion in Cell Biology. 25 (1): 134–141. doi:10.1016/j.ceb.2012.11.005. PMC 4927265. PMID 23312067.
- ^ a b Cavalier-Smith, Thomas; Chao, Ema E.; Lewis, Rhodri (2016-06-01). "187-gene phylogeny of protozoan phylum Amoebozoa reveals a new class (Cutosea) of deep-branching, ultrastructurally unique, enveloped marine Lobosa and clarifies amoeba evolution". Molecular Phylogenetics and Evolution. 99: 275–296. doi:10.1016/j.ympev.2016.03.023.
- ^ Simpson AG (2003). "Cytoskeletal organization, phylogenetic affinities and systematics in the contentious taxon Excavata (Eukaryota)". Int. J. Syst. Evol. Microbiol. 53 (Pt 6): 1759–77. doi:10.1099/ijs.0.02578-0. PMID 14657103.
- ^ Naiara Rodríguez-Ezpeleta, Henner Brinkmann, Gertraud Burger, Andrew J. Roger, Michael W. Gray, Hervé Philippe, and B. Franz Lang (2007). "Toward Resolving the Eukaryotic Tree: The Phylogenetic Positions of Jakobids and Cercozoans". Curr. Biol. 17 (16): 1420–1425. doi:10.1016/j.cub.2007.07.036. PMID 17689961.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Laura Wegener Parfrey; Erika Barbero; Elyse Lasser; Micah Dunthorn; Debashish Bhattacharya; David J Patterson; Laura A Katz (2006). "Evaluating Support for the Current Classification of Eukaryotic Diversity". PLoS Genet. 2 (12): e220. doi:10.1371/journal.pgen.0020220. PMC 1713255. PMID 17194223.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Zaremba-Niedzwiedzka, Katarzyna; Caceres, Eva F.; Saw, Jimmy H.; Bäckström, Disa; Juzokaite, Lina; Vancaester, Emmelien; Seitz, Kiley W.; Anantharaman, Karthik; Starnawski, Piotr (2017-01-11). "Asgard archaea illuminate the origin of eukaryotic cellular complexity". Nature. advance online publication. doi:10.1038/nature21031. ISSN 1476-4687.
- ^ Derelle, Romain; Torruella, Guifré; Klimeš, Vladimír; Brinkmann, Henner; Kim, Eunsoo; Vlček, Čestmír; Lang, B. Franz; Eliáš, Marek (2015-02-17). "Bacterial proteins pinpoint a single eukaryotic root". Proceedings of the National Academy of Sciences. 112 (7): E693–E699. doi:10.1073/pnas.1420657112. ISSN 0027-8424. PMC 4343179. PMID 25646484.
- ^ Cavalier-Smith, T.; Chao, E. E.; Snell, E. A.; Berney, C.; Fiore-Donno, A. M.; Lewis, R. (2014). "Multigene eukaryote phylogeny reveals the likely protozoan ancestors of opisthokonts (animals, fungi, choanozoans) and Amoebozoa". Molecular Phylogenetics & Evolution. 81: 71–85. doi:10.1016/j.ympev.2014.08.012.
- ^ Cavalier-Smith, Thomas (2010-06-23). "Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree". Biology Letters. 6 (3): 342–345. doi:10.1098/rsbl.2009.0948. ISSN 1744-9561. PMC 2880060. PMID 20031978.
- ^ He, Ding; Fiz-Palacios, Omar; Fu, Cheng-Jie; Fehling, Johanna; Tsai, Chun-Chieh; Baldauf, Sandra L. "An Alternative Root for the Eukaryote Tree of Life". Current Biology. 24 (4): 465–470. doi:10.1016/j.cub.2014.01.036.
- ^ Cavelier Smith. "Early evolution of eukaryote ...". Elsevier. doi:10.1016/j.ejop.2012.06.001.
- ^ Hug, Laura A.; Baker, Brett J.; Anantharaman, Karthik; Brown, Christopher T.; Probst, Alexander J.; Castelle, Cindy J.; Butterfield, Cristina N.; Hernsdorf, Alex W.; Amano, Yuki (2016-04-11). "A new view of the tree of life". Nature Microbiology. 1 (5). doi:10.1038/nmicrobiol.2016.48. ISSN 2058-5276.
- ^ Cavalier-Smith, Thomas (2016-10-01). "Higher classification and phylogeny of Euglenozoa". European Journal of Protistology. 56: 250–276. doi:10.1016/j.ejop.2016.09.003.
- ^ Derelle, Romain; Torruella, Guifré; Klimeš, Vladimír; Brinkmann, Henner; Kim, Eunsoo; Vlček, Čestmír; Lang, B. Franz; Eliáš, Marek (2015-02-17). "Bacterial proteins pinpoint a single eukaryotic root". Proceedings of the National Academy of Sciences. 112 (7): E693–E699. doi:10.1073/pnas.1420657112. ISSN 0027-8424. PMC 4343179. PMID 25646484.
{{cite journal}}: CS1 maint: PMC format (link)