| bZIP transcription factor | |||||||||
|---|---|---|---|---|---|---|---|---|---|
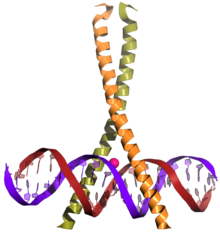 CREB (top) is a transcription factor capable of binding DNA via the bZIP domain (bottom) and regulating gene expression. | |||||||||
| Identifiers | |||||||||
| Symbol | bZIP_1 | ||||||||
| Pfam | PF00170 | ||||||||
| InterPro | IPR011616 | ||||||||
| PROSITE | PDOC00036 | ||||||||
| SCOP2 | 1ysa / SCOPe / SUPFAM | ||||||||
| CDD | cd14686 | ||||||||
| Membranome | 235 | ||||||||
| |||||||||
The Basic Leucine Zipper Domain (bZIP domain) is found in many DNA binding eukaryotic proteins. One part of the domain contains a region that mediates sequence specific DNA binding properties and the leucine zipper that is required to hold together (dimerize) two DNA binding regions. The DNA binding region comprises a number of basic amino acids such as arginine and lysine. Proteins containing this domain are transcription factors.[1][2]
bZIP transcription factors[edit]
bZIP transcription factors are found in all eukaryotes and form one of the largest families of dimerizing TFs.[3] An evolutionary study from 2008 revealed that 4 bZIP genes were encoded by the genome of the most recent common ancestor of all plants.[4] Interactions between bZIP transcription factors are numerous and complex [5][6][3] and play important roles in cancer development[7] in epithelial tissues, steroid hormone synthesis by cells of endocrine tissues,[8] factors affecting reproductive functions,[9] and several other phenomena that affect human health.
bZIP domain containing proteins[edit]
- AP-1 fos/jun heterodimer that forms a transcription factor
- Jun-B transcription factor
- CREB cAMP response element transcription factor
- OPAQUE2 (O2) transcription factor of the 22-kD zein gene that encodes a class of storage proteins in the endosperm of maize (Zea mays) kernels
- NFE2L2 or Nrf2
- Bzip Maf transcription factors
Human proteins containing this domain[edit]
ATF1; ATF2; ATF4; ATF5; ATF6; ATF7; BACH1; BACH2; BATF; BATF2; CEBPA; CEBPB; CEBPD; CEBPE; CEBPG; CEBPZ; CREB1; CREB3; CREB3L1; CREB3L2; CREB3L3; CREB3L4; CREB5; CREBL1; CREM; E4BP4; FOSL1; FOSL2; JUN; JUNB; JUND; MAFA; MAFB; MAFF; MAFG; NRL; C-MAF; MAFK; NFE2; NFE2L2; NFE2L3; SNFT; XBP1
References[edit]
- ^ Ellenberger T (1994). "Getting a grip in DNA recognition: structures of the basic region leucine zipper, and the basic region helix-loop-helix DNA-binding domains". Curr. Opin. Struct. Biol. 4 (1): 12–21. doi:10.1016/S0959-440X(94)90054-X.
- ^ Hurst HC (1995). "Transcription factors 1: bZIP proteins". Protein Profile. 2 (2): 101–68. PMID 7780801.
- ^ a b Amoutzias, Grigoris D.; Robertson, David L.; Van de Peer, Yves; Oliver, Stephen G. (2008-05-01). "Choose your partners: dimerization in eukaryotic transcription factors". Trends in Biochemical Sciences. 33 (5): 220–229. doi:10.1016/j.tibs.2008.02.002. ISSN 0968-0004. PMID 18406148.
- ^ Corrêa LG, Riaño-Pachón DM, Schrago CG, dos Santos RV, Mueller-Roeber B, Vincentz M (2008). Shiu SH (ed.). "The Role of bZIP Transcription Factors in Green Plant Evolution: Adaptive Features Emerging from Four Founder Genes". PLOS ONE. 3 (8): e2944. Bibcode:2008PLoSO...3.2944C. doi:10.1371/journal.pone.0002944. PMC 2492810. PMID 18698409.
- ^ Vinson, Charles; Acharya, Asha; Taparowsky, Elizabeth J. (2006-01-01). "Deciphering B-ZIP transcription factor interactions in vitro and in vivo". Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression. 1759 (1–2): 4–12. doi:10.1016/j.bbaexp.2005.12.005. ISSN 0006-3002. PMID 16580748.
- ^ Newman, John R. S.; Keating, Amy E. (2003-06-27). "Comprehensive identification of human bZIP interactions with coiled-coil arrays". Science. 300 (5628): 2097–2101. Bibcode:2003Sci...300.2097N. doi:10.1126/science.1084648. ISSN 1095-9203. PMID 12805554. S2CID 36715183.
- ^ Vlahopoulos SA, Logotheti S, Mikas D, Giarika A, Gorgoulis V, Zoumpourlis V (April 2008). "The role of ATF-2 in oncogenesis". BioEssays. 30 (4): 314–27. doi:10.1002/bies.20734. PMID 18348191. S2CID 678541.
- ^ Manna PR, Dyson MT, Eubank DW, Clark BJ, Lalli E, Sassone-Corsi P, Zeleznik AJ, Stocco DM (January 2002). "Regulation of steroidogenesis and the steroidogenic acute regulatory protein by a member of the cAMP response-element binding protein family". Mol. Endocrinol. 16 (1): 184–99. doi:10.1210/mend.16.1.0759. PMID 11773448.
- ^ Hoare S, Copland JA, Wood TG, Jeng YJ, Izban MG, Soloff MS (May 1999). "Identification of a GABP alpha/beta binding site involved in the induction of oxytocin receptor gene expression in human breast cells, potentiation by c-Fos/c-Jun". Endocrinology. 140 (5): 2268–79. doi:10.1210/endo.140.5.6710. PMID 10218980.
Well, that’s interesting to know that Psilotum nudum are known as whisk ferns. Psilotum nudum is the commoner species of the two. While the P. flaccidum is a rare species and is found in the tropical islands. Both the species are usually epiphytic in habit and grow upon tree ferns. These species may also be terrestrial and grow in humus or in the crevices of the rocks.
View the detailed Guide of Psilotum nudum: Detailed Study Of Psilotum Nudum (Whisk Fern), Classification, Anatomy, Reproduction