| Terrabacteria | |
|---|---|
| |
| Scanning electron micrograph of Actinomyces israelii (Actinomycetota) | |
| Scientific classification | |
| Domain: | Bacteria |
| Clade: | Terrabacteria Battistuzzi et al., 2004, Battistuzzi & Hedges, 2009 |
| Phyla | |
| Synonyms | |
| |
Terrabacteria is a taxon containing approximately two-thirds of prokaryote species, including those in the gram positive phyla (Actinomycetota and Bacillota) as well as the phyla "Cyanobacteria", Chloroflexota, and Deinococcota.[1][2]
It derives its name (terra = "land") from the evolutionary pressures of life on land. Terrabacteria possess important adaptations such as resistance to environmental hazards (e.g., desiccation, ultraviolet radiation, and high salinity) and oxygenic photosynthesis. Also, the unique properties of the cell wall in gram-positive taxa, which likely evolved in response to terrestrial conditions, have contributed toward pathogenicity in many species.[2] These results now leave open the possibility that terrestrial adaptations may have played a larger role in prokaryote evolution than currently understood.[1][2]
Terrabacteria was proposed in 2004 for Actinomycetota, "Cyanobacteria", and Deinococcota[1] and was expanded later to include Bacillota and Chloroflexota.[2] Other phylogenetic analyses [3][4][5] have supported the close relationships of these phyla. Most species of prokaryotes not placed in Terrabacteria were assigned to the taxon Hydrobacteria,[2] in reference to the moist environment inferred for the common ancestor of those species. Some molecular phylogenetic analyses [6][7] have not supported this dichotomy of Terrabacteria and Hydrobacteria, but the most recent genomic analyses,[4][5] including those that have focused on rooting the tree,[4] have found these two groups to be monophyletic.[4]
Terrabacteria and Hydrobacteria were inferred to have diverged approximately 3 billion years ago, suggesting that land (continents) had been colonized by prokaryotes at that time.[2] Together, Terrabacteria and Hydrobacteria form a large group containing 97% of prokaryotes and 99% of all species of Bacteria known by 2009, and placed in the taxon Selabacteria, in allusion to their phototrophic abilities (selas = light).[8] Currently, the bacterial phyla that are outside of Terrabacteria + Hydrobacteria, and thus justifying the taxon Selabacteria, are debated and may or may not include Fusobacteria.[2][4]
The name “Glidobacteria” [9] included some members of Terrabacteria but excluded the large gram positive groups, Bacillota and Actinomycetota, and is not supported by molecular phylogenetic data.[1][2][3][6][7][4][5] Moreover, the article naming Glidobacteria [9] did not include a molecular phylogeny or statistical analyses and did not follow the widely used three-domain system. For example, it claimed that eukaryotes split from Archaea very recently (~900 Mya), which is contradicted by the fossil record,[10] and that lineage of eukaryotes + Archaea was nested within Bacteria as a close relative of Actinomycetota.
In 2022, new rules were introduced for kingdom-level taxa of prokaryotes, and the same two authors who proposed those new rules, proposed new names in 2024.[11] They concluded that “the taxonomically preferable solution for bacterial kingdoms seems to be to accept the subdivision apparent in the study by Battistuzzi and Hedges [2],” with refinement. However, instead of using the existing names Terrabacteria and Hydrobacteria, or those names with modified (kingdom-level) endings, Terrabacteriati and Hydrobacteriati, they coined their own new names, Bacillati and Pseudomonadati, respectively. As an action that creates more instability than stability, it is likely to be challenged.
Phylogeny[edit]
The phylogenetic tree according to the phylogenetic analyses of Battistuzzi and Hedges (2009) is the following and with a molecular clock calibration.[1][2]
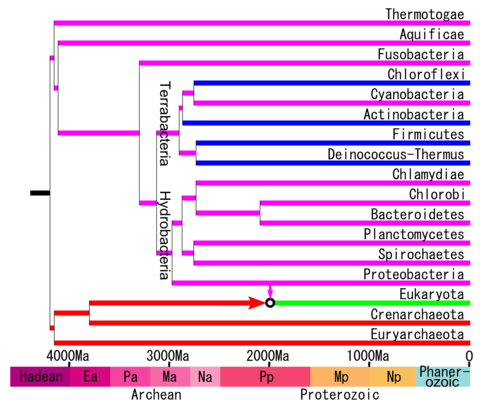
Recent molecular analyses have found roughly the following relationships including other phyla, whose relationships were uncertain.[12][13][14][15][16][17]
| Terrabacteria |
| ||||||||||||||||||||||||||||||
On the other hand, Coleman et al.[4] named the clade composed of Thermotogota, Deinococcota, Synergistota and related as DST and furthermore the analysis suggests that ultra-small bacteria (CPR group) may belong to Terrabacteria being more closely related to Chloroflexota. According to this study the phylum Aquificota sometimes included belongs to Hydrobacteria and that the phylum Fusobacteriota can belong to both Terrabacteria and Hydrobacteria. The result was the following:[4]
| Terrabacteria | |
References[edit]
- ^ a b c d e Battistuzzi FU, Feijao A, Hedges SB (November 2004). "A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land". BMC Evolutionary Biology. 4: 44. doi:10.1186/1471-2148-4-44. PMC 533871. PMID 15535883.
- ^ a b c d e f g h i j Battistuzzi FU, Hedges SB (February 2009). "A major clade of prokaryotes with ancient adaptations to life on land". Molecular Biology and Evolution. 26 (2): 335–343. doi:10.1093/molbev/msn247. PMID 18988685.
- ^ a b Bern M, Goldberg D (May 2005). "Automatic selection of representative proteins for bacterial phylogeny". BMC Evolutionary Biology. 5 (1): 34. doi:10.1186/1471-2148-5-34. PMC 1175084. PMID 15927057.
- ^ a b c d e f g h Coleman GA, Davín AA, Mahendrarajah TA, Szánthó LL, Spang A, Hugenholtz P, et al. (May 2021). "A rooted phylogeny resolves early bacterial evolution". Science. 372 (6542): eabe0511. doi:10.1126/science.abe0511. hdl:1983/51e9e402-36b7-47a6-91de-32b8cf7320d2. PMID 33958449. S2CID 233872903.
- ^ a b c Léonard RR, Sauvage E, Lupo V, Perrin A, Sirjacobs D, Charlier P, et al. (February 2022). "Was the Last Bacterial Common Ancestor a Monoderm after All?". Genes. 13 (2): 376. doi:10.3390/genes13020376. PMC 8871954. PMID 35205421.
- ^ a b Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, et al. (April 2016). "A new view of the tree of life". Nature Microbiology. 1 (5): 16048. doi:10.1038/nmicrobiol.2016.48. PMID 27572647. S2CID 3833474.
- ^ a b Zhu Q, Mai U, Pfeiffer W, Janssen S, Asnicar F, Sanders JG, et al. (December 2019). "Phylogenomics of 10,575 genomes reveals evolutionary proximity between domains Bacteria and Archaea". Nature Communications. 10 (1): 5477. Bibcode:2019NatCo..10.5477Z. doi:10.1038/s41467-019-13443-4. PMC 6889312. PMID 31792218.
- ^ Battistuzzi FU, Hedges SB (2009). "Eubacteria". In Hedges SB, Kumar S (eds.). The Timetree of Life. New York: Oxford University Press. pp. 106–115.
- ^ a b Cavalier-Smith T (July 2006). "Rooting the tree of life by transition analyses". Biology Direct. 1 (1): 19. doi:10.1186/1745-6150-1-19. PMC 1586193. PMID 16834776.
- ^ Knoll AH (2003). Life on a Young Planet : The First Three Billion Years of Evolution on Earth - Updated Edition. ISBN 0-691-00978-3. OCLC 1303471348.
- ^ Göker, Markus; Oren, Aharon (2024-01-22). "Valid publication of names of two domains and seven kingdoms of prokaryotes". International Journal of Systematic and Evolutionary Microbiology. 74 (1). doi:10.1099/ijsem.0.006242. ISSN 1466-5026.
- ^ Anantharaman K, Brown CT, Hug LA, Sharon I, Castelle CJ, Probst AJ, et al. (October 2016). "Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system". Nature Communications. 7: 13219. Bibcode:2016NatCo...713219A. doi:10.1038/ncomms13219. PMC 5079060. PMID 27774985.
- ^ Matheus Carnevali PB, Schulz F, Castelle CJ, Kantor RS, Shih PM, Sharon I, et al. (January 2019). "Hydrogen-based metabolism as an ancestral trait in lineages sibling to the Cyanobacteria". Nature Communications. 10 (1): 463. Bibcode:2019NatCo..10..463M. doi:10.1038/s41467-018-08246-y. PMC 6349859. PMID 30692531.
- ^ Ji M, Greening C, Vanwonterghem I, Carere CR, Bay SK, Steen JA, et al. (December 2017). "Atmospheric trace gases support primary production in Antarctic desert surface soil". Nature. 552 (7685): 400–403. Bibcode:2017Natur.552..400J. doi:10.1038/nature25014. hdl:2440/124244. PMID 29211716. S2CID 4394421.
- ^ Tahon G, Tytgat B, Lebbe L, Carlier A, Willems A (July 2018). "Abditibacterium utsteinense sp. nov., the first cultivated member of candidate phylum FBP, isolated from ice-free Antarctic soil samples". Systematic and Applied Microbiology. 41 (4): 279–290. doi:10.1016/j.syapm.2018.01.009. PMID 29475572. S2CID 3515091.
- ^ Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, et al. (July 2013). "Insights into the phylogeny and coding potential of microbial dark matter". Nature. 499 (7459): 431–437. Bibcode:2013Natur.499..431R. doi:10.1038/nature12352. hdl:10453/27467. PMID 23851394. S2CID 4394530.
- ^ Eloe-Fadrosh EA, Paez-Espino D, Jarett J, Dunfield PF, Hedlund BP, Dekas AE, et al. (January 2016). "Global metagenomic survey reveals a new bacterial candidate phylum in geothermal springs". Nature Communications. 7: 10476. Bibcode:2016NatCo...710476E. doi:10.1038/ncomms10476. PMC 4737851. PMID 26814032.