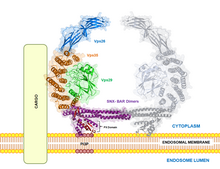
Vacuolar protein sorting ortholog 35 (VPS35) is a protein involved in autophagy and is implicated in neurodegenerative diseases, such as Parkinson's disease (PD) and Alzheimer's disease (AD).[1][2][3][4][5] VPS35 is part of a complex called the retromer, which is responsible for transporting select cargo proteins between vesicular structures (e.g., endosomes, lysosomes, vacuoles) and the Golgi apparatus.[1][6][7][8][9] Mutations in the VPS35 gene (VPS35) cause aberrant autophagy, where cargo proteins fail to be transported and dysfunctional or unnecessary proteins fail to be degraded.[5][7] There are numerous pathways affected by altered VPS35 levels and activity, which have clinical significance in neurodegeneration.[1][2][3][4][5] There is therapeutic relevance for VPS35, as interventions aimed at correcting VPS35 function are in speculation.[5][10][11]
| VPS35 | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | VPS35, MEM3, PARK17, VPS35 retromer complex component, retromer complex component | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 601501 MGI: 1890467 HomoloGene: 6221 GeneCards: VPS35 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Gene[edit]
In humans, VPS35 is transcribed on chromosome 16q11.2 where is spans about 29.6 kilobases and contains 17 exons.[10][11][16][17] It is evolutionarily conserved and required for survival, as mouse knockout studies have demonstrated embryonic lethality.[2][3][6][8][9][10][11] VPS35 levels peak at postnatal days 10-15 and then decline to a low, stable level throughout adulthood.[3] RNA expression of VPS35 is ubiquitous throughout the body, but are higher in the brain, heart, gonads, spleen, and skeletal muscle, and lower in the lung, liver, kidney, and blood leukocytes.[16][17]
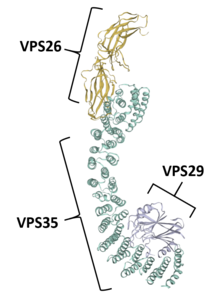
Protein[edit]
VPS35 was first identified in Saccharomyces cerevisiae from a study investigating the formation of lysosome-like vacuoles and sorting of vacuolar proteins.[4][5][17] The protein contains 796 amino acid residues, with a molecular mass of 92 kDa and an isoelectric point of 5.32.[11][16][17]
Structure[edit]
VPS35 binds with other proteins to form the retromer, an evolutionarily conserved complex that plays a major role in transmembrane protein recycling from endosomes to the trans-Golgi network (TGN).[1][6][7][8][9] VPS35 itself folds into a secondary structure that represents an α-helical solenoid, containing 34 α-helix repeats.[17]
As part of the retromer, VPS35 trimerizes with other vacuolar protein sorting orthologs, VPS26 and VPS29. In less common situations, VPS35 can bind VPS26 and VPS29 alone, creating heterodimers.[5] VPS26 binds the N-terminus of VPS35 at a conserved PRLYL motif (residues 1-172), whereas a C-terminal α-solenoid fold (residues 307-796) binds VPS29.[1][16][17] These VPS orthologs stabilize each other within the retromer; VPS35 knockdown can lead to VPS29 degradation, and vice versa.[5][7] The VPS35, VPS26, and VPS29 trimer forms the cargo recognition complex, a necessary component for the retromer's ability to regulate vesicular sorting.[1][2][3][5][6][7][8][9][10][18] This is achieved by specifically targeting sorting nexins (i.e., SNX1, SNX2, SNX5, SNX6, and SNX32), which anchor the retromer to endosomes and other vesicular structures.[3][11][17][18]
Function[edit]

After translation, VPS35 localizes to the endosome, where it, along with the retromer, has been studied in various phagocytotic processes.[4][10][17] Sortilin and the cation-independent mannose-6-phosphate receptor (CI-MPR) regulate lysosome-mediated degradation and are known VPS35 cargo-proteins.[10] The Wiskott Aldrich Syndrome protein and scar homologue (WASH) complex, directly recruited by VPS35, regulates macroautophagy, whereby an autophagosome engulfs select proteins and whole organelles for lysosomal degradation.[10][19] VPS35 also recruits lysosome-associated membrane protein 2a (LAMP2a) to help initiate chaperone-mediated autophagy, whereby a heat shock protein 70kDa protein 8 (HSPA8)-containing complex facilitates degradation of proteins containing a KFERQ signal.[10][19]
Mutations in VPS35 lead to generalized endosomal defects, such as vesicle enlargement and sequestration around the nucleus.[5] These mutations also affect many cellular processes overseen by the retromer complex, including vesicular trafficking, plasma membrane receptor recycling, protein aggregation, mitochondrial function, and dopamine signaling.[7] These processes play a role in neurodegenerative diseases.[1][2][3][4][5]
Clinical significance[edit]
Parkinson's disease[edit]
PD is a clinical challenge, presenting itself as the second most common neurodegenerative disease.[11][20] It is characterized by dopaminergic neuron loss in the substantia nigra pars compacta and the toxic aggregation of the α-synuclein protein into Lewy Bodies, all of which cause motor dysfunctions and dopamine deficiency in PD patients.[11][20] VPS35 mutations and dysfunctional retromer-mediated protein sorting and recycling are implicated in neurodegenerative processes relevant to PD.[8]
Genetic variants[edit]
In 2011, exome sequencing was performed on a Swiss family where all six members developed late-onset, autosomal dominant PD.[3][5][9][11][20][21][22] The sequencing revealed a mutation from an aspartic acid to asparagine at amino acid residue 620 (D620N) in VPS35.[8][9] Sequence alignment of VPS35 orthologs between Homo sapiens, Pan troglodytes, Mus musculus, Rattus norvegicus, Bos taurus, Canis familiaris, Gallus gallus, Xenopus laevis, Danio rerio, Drosophila melanogaster, and Saccharomyces cerevisiae has shown that the 620 position is highly mutagenic with a propensity to substitute aspartic acid with asparagine throughout evolution.[8] Geographically, the VPS35-D620N variant is most prevalent among Caucasian descendants and families in Switzerland, Austria, the United States, Tunisia, Israel, the United Kingdom, France, Germany, and Japan.[2][9][10][22] Other VPS35 variants (e.g., P316S, R32S, R524W, I560T, H599R, M607V) have been identified among some PD patients; however, the VPS35-D620N variant has been the most extensively characterized and is currently the only mutated form of VPS35 confirmed to be pathogenic.[1][7][9] It is currently unknown how frequently the D620N mutation appears in the general population.[17]
Patient characteristics[edit]
The neurochemistry and clinical presentation between VPS35-induced PD and idiopathic PD are not significantly different.[9][11][21] PD patients with VPS35 mutations experience typical PD symptoms: bradykinesia (91.4%), rigidity (80%), tremor (77.1%), and postural instability (60%).[9][11][16][21] Patients with the VPS35-D620N variant experience a mean age of disease onset between 50–52 years (earlier than classic PD onset of 65–85 years), are typically white (82.9%), and have high familial history (91.4%).[8][10][11][16][21] The VPS35-D620N mutation occurs more frequently in familial PD (~1.3%) than in sporadic PD (0.3%).[11] PD patients harboring the VPS35-D620N mutation follow slow disease progression; cognitive or neuropsychiatric aspects are generally spared.[23] All patients respond well to levodopa therapy, one of the main treatments to alleviate PD symptoms.[9][11] Overall, the VPS35-D620N mutation is relatively rare in PD, with estimated prevalence of 0.115% from over a dozen case studies comprising approximately 22,000 PD patients globally.[9][17] One clinical caveat is that Lewy body pathology has not been fully understood in the context of VPS35-induced PD, as only a single PD patient harboring the VPS35-D620N variant has undergone an autopsy.[9] However, it is known that in brains of sporadic PD patients, there is mislocalized VPS35 in Lewy bodies.[10]
Interactions between VPS35 and PD-linked genes[edit]
Changes in VPS35 affect levels of leucine-rich repeat kinase 2 (LRRK2), a candidate gene involved in PD that helps vesicular trafficking by phosphorylating Rab proteins.[5][10][17][24] VPS35 knockdown or overexpression of the VPS35-D620N variant can enhance autophosphorylation of LRRK2, increasing its overall activity.[17] Transgenic mice harboring the VPS35-D620N variant have demonstrated altered Rab phosphorylation by LRRK2.[10] In Drosophila, overexpression of VPS35 can rescue mutant LRRK2-induced PD phenotypes.[9][17] The LRRK2 mutation, G2019S, has also been found to decrease VPS35 levels in mouse N2A neuroblastoma cells, indicating an affective mechanism between these factors.[10]
Parkin, an E3 ubiquitin ligase involved in protein degradation, is commonly seen in autosomal recessive juvenile parkinsonism and has known interactions with VPS35.[17] VPS35 overexpression in Drosophila lacking Parkin reverses Parkin-deficient phenotypes, increasing longevity, climbing ability, and decreasing sensitivity to paraquat, a toxic herbicide known to be associated with PD onset in humans.[17] Parkin is known to increase ubiquitination of VPS35.[17] Counterintuitively, this does not lead to proteasomal degradation of VPS35; a functional role is still being investigated.[24] It is proposed that VPS35 may act downstream of Parkin, since overexpression of VPS35-D620N in Parkin knockout mice has not been shown to rescue dopaminergic neuron loss.[17]
Alzheimer's disease[edit]
AD is the most prominent cause of dementia (60-80%) and affects many cognitive abilities in patients, including word retrieval, memory recall, and other general executive functions necessary for basic self-care.[25] AD pathology typically begins with the formation of amyloid beta (Aβ) plaques in the brain, which trigger an inflammatory response by microglia and cause a cascade of tau accumulation and spreading.[26] These changes result in the degeneration of neurons, leading to a loss of synaptic connections and neurotransmitter signaling.[25][26]
Clinically, low expression of VPS35 in the brain is a risk factor of AD, since it is known that regions high in AD pathology show low VPS35 activity.[5][6][7][27] Specifically, there are decreased levels of VPS35 in the hippocampi of postmortem AD patients relative to healthy patients.[2][7][8][16] This can be modeled in a specific strain of AD-like mice, Tg2576, whereby heterozygous deletion of VPS35 enhances AD phenotypes in the hippocampus and cortex.[9] Humans possessing a VPS35 genetic variant also increases the risk of developing AD.[8] Consequently, AD pathology is linked to aberrant retromer function.[11]
VPS35 knockdown studies have demonstrated increased amyloid precursor protein (APP) and Aβ plaque levels, hallmarks of AD.[3] VPS35 insufficiency reduces transport of endosomes containing APP, ultimately facilitating APP aggregation and formation of Aβ plaques.[1][5][11] Sortilin-related receptor is a cargo protein that interacts with VPS35; it binds APP and delivers APP to the lysosomal system for degradation.[6] Impairment of retromer activity by VPS35 mutation also increases beta-secretase 1 (BACE1) activity, which cleaves larger APPs and enhances Aβ plaque toxicity.[2][3] This is observed in heterozygous mice deficient in VPS35, which have greater amounts of Aβ40 and Aβ42 compared to controls.[1] Decreased expression of VPS35 in Drosophila AD models further shows an increase in Aβ plaque formation, BACE1 activity, memory deficits, and synaptic dysfunction.[6][7][16]
VPS35 deletion in mammalian models of AD is associated with aberrant microglia function and impaired hippocampal development; however, causal variants have yet to be determined.[7] Possibly, VPS35 loss-of-function may impair an inflammatory response in AD, since triggering receptor expressed on myeloid cells 2 (TREM2), a microglial factor associated with inflammation in AD, is a cargo protein of the retromer and VPS35.[17] Tau pathology is also observed in the brains of VPS35-D620N knock-in mice.[10][11]
Pathophysiology[edit]
Aberrant autophagy[edit]
The WASH complex is an important component in autophagy and endosome function.[10] It helps form actin patches on endosomal membranes to facilitate their transport to the TGN.[9] The C-terminus of VPS35 binds the WASH complex; through the retromer, this interaction helps distinguish which proteins are retrieved from the TGN.[2][9][17] Compared to non-mutated VPS35, the VPS35-D620N variant has weaker binding affinity to family with sequence similarity 210 member A (FAM21), a component of the WASH complex, leading to autophagic dysfunction.[9][11] A further consequence of impaired WASH complex binding is improper trafficking of autophagy-related protein 9a (ATG9A), a transmembrane protein that facilitates the interaction between LC3 and autophagosomes.[1][11] Disruption in ATG9A trafficking by the VPS35-D620N mutation also decreases WASH complex localization to the endosomes, suggesting a cyclic feedback between mutated VPS35 and ATG9A mislocalization.[3][10][11] Another result of impaired WASH complex binding is the missorting of Cl-MPR and glucose transporter 1, GLUT1, affecting TGN function and energy utilization.[9]
In PD, specifically, iron accumulation can alter the retromer's function in autophagy. Relative to healthy patients, higher amounts of iron are present in the substantia nigra of PD patients, as well as divalent metal transporter 1 (DMT1), a retromer-associated protein that sequesters iron in cells.[16][17] It is thought that the combination of heightened iron concentration and DMT1 activity leads to neuronal death in PD by increasing α-synuclein accumulation.[16] SH-SY5Y cells expressing the VPS35-D620N mutant display DMT1 accumulation.[17] VPS35 downregulation by RNAi allows DMT1 to associate more closely with LAMP2-positive structures, increasing iron concentration in lysosomes and impairing autophagy.[16] This effect is reversed with restoration of VPS35.[16] Holistically, chaperone-mediated autophagy is disrupted with impaired WASH complex function; heterozygous-VPS35 knockout mice and mice with the VPS35-D620N mutation have a reduction in LAMP2a trafficking, which leads to α-synuclein accumulation in the brain and poses an important functional implication in PD.[9][11]

Α-synuclein accumulation and dopaminergic neuron loss[edit]
Deficient VPS35 expression or bearing the VPS35-D620N variant is known to cause α-synuclein aggregation and dopaminergic neuron loss, relevant characteristics in PD.[3][7][10][11][17][20] This has been studied in Drosophila, where VPS35 deficiency leads to α-synuclein accumulation in lysosomes and inappropriate trafficking of CI-MPR and its ligand, cathepsin D, a lysosomal protease that is important for α-synuclein processing.[9][11][20] Knockdown of VPS35 using short hairpin RNA decreases dopamine transporter localization and recycling at the synaptic membrane.[7][8][11] Mouse heterozygous knockouts of VPS35 display PD-like phenotypes into adulthood, including α-synuclein aggregation, motor impairments, and a loss of dopaminergic activity in the substantia nigra and striatum.[9][11] Transgenic mice overexpressing wildtype human α-synuclein have lower levels of VPS35, and viral overexpression of wildtype VPS35, but not mutant VPS35, can restore hippocampal neuronal loss and reduce astrogliosis.[9][11]
It is unclear in animal studies whether the VPS35-D620N variant is sufficient to cause PD pathology. In rats, overexpression by adeno-associated virus (AAV) of human VPS35-D620N does not alter α-synuclein levels, phosphorylation, or PD pathology in dopaminergic neurons in the substantia nigra.[9] However, other studies using rats have shown that introducing the VPS35-D620N variant results in dopaminergic neuronal degeneration in the substantia nigra.[1] Mouse knock-in models of VPS35-D620N display no difference in retromer assembly and stability or protein levels related to endolysosomes, autophagy, mitochondria, or α-synuclein in the brain, although there is a reduction in striatal dopamine around 5 months of age.[9][11] In Drosophila, overexpression of human VPS35-D620N or VPS35-P316S variants shows dopaminergic neuron loss, locomotor impairments, and decreased overall survival relative to human wildtype VPS35.[9]
The Wnt/β-catenin (or Wnt-PCP) pathway may participate in the degeneration of dopaminergic neurons in the midbrain.[16] Inactivity of the retromer complex through VPS35 mutation is thought to degrade Wntless, a membrane protein that regulates Wnt secretion from cells.[16] This leads to downregulated Wnt signaling, increasing vulnerability and degradation of dopaminergic neurons.[16]
Impaired glutamatergic signaling[edit]
Loss of VPS35 activity decreases signaling efficiency of the excitatory neurotransmitter, glutamate.[17] This has also been shown using a neuron-specific mutation of VPS35-D620N.[17] Mechanistically, this may be mediated by the retromer, where it has been shown to localize to dendritic spines and mediate turnover of the glutamatergic AMPA receptor, GluR1.[9][17] Because of impaired WASH complex binding through the VPS35-D620N mutation, GluR1 can be mistrafficked.[11] Treating mouse hippocampal and cortical neurons with VPS35 small interfering RNA inhibits AMPA receptor trafficking to the dendritic membrane.[7] VPS35-heterozygous knockout mice show similar effects, with an additional observation of impaired dendritic spine development.[9] Human induced pluripotent stem cell-derived dopaminergic neurons with the VPS35-D620N mutation also shift GluR1 localization away from dendritic spines, altering glutamatergic synaptic transmission.[9]
AMPA receptor localization is important for synaptic plasticity, because knockdown of VPS35 in the hippocampus blocks long term potentiation.[3] However, overall synaptic transmission is not profoundly affected.[3] Current knowledge suggests that alteration in VPS35 levels only affects excitatory neuronal signaling, because inhibitory GABA receptors do not appear to be affected by VPS35 or retromer impairment.[3]
Disrupted mitochondrial homeostasis[edit]
Mitochondria are organelles that undergo oxidative phosphorylation to produce adenosine triphosphate, or ATP, providing energy for the cell to carry out necessary metabolic and homeostatic processes.[10] VPS35 and the retromer help create mitochondria-derived vesicles (MDVs), direct mitochondrial proteins toward degradation as needed, and facilitate cross-talk between vesicles and peroxisomes or lysosomes.[9][17] In disease, deficient or mutated VPS35 causes morphological and functional abnormalities of mitochondria.[10] Mutated VPS35 can lead to mitochondrial membrane instability by ceramide accumulation, which promotes the production of reactive oxygen species.[10] This lowers mitochondrial membrane potential, decreases production of ATP, and impairs bioenergetics.[9] Additionally, some MDVs can be positive for mitochondrial-anchored protein ligase (MAPL), which helps the retromer transport MDVs to peroxisomes for oxidation.[20] Knockdown of VPS35 interferes with this transport.[20]
VPS35 and the retromer also help modulate mitochondrial fusion and fission.[10][11][17] In healthy conditions, VPS35 helps regulate mitochondrial fusion by removing mitochondrial E3 ubiquitin protein ligase 1 (MUL1) from the outer mitochondrial membrane and preventing the degradation of mitofusin 2 (Mfn2).[10] VPS35 loss-of-function increases MUL1 in dopaminergic neurons, leading to ubiquitination and degradation of Mfn2.[10] Treating VPS35-deficient neurons with wildtype VPS35, but not the VPS35-D620N mutant, restores MUL1 levels, and decreases mitochondrial fragmentation.[9] Instead, overexpression of the human VPS35-D620N mutant in cultured rat cortical neurons, M17 cells, and human fibroblasts, and mouse substantia nigral neurons in vivo, causes mitochondrial fragmentation and neuronal loss, relative to a different VPS35 variant, R524W.[9] VPS35 loss-of-function also facilitates mitochondrial fission through the retention of inactive dynamin related protein 1 (Drp1) in the outer mitochondrial membrane.[10]
Therapeutic application[edit]
Thiophene thiourea derivatives R33 and R55 have been shown to regulate VPS35 levels, restabilizing the retromer complex and reestablishing endosomal function in AD.[10] Rapamycin treatment has also been shown to enhance autophagic function and improve clearance of protein aggregates that pay key roles in PD and AD.[11]
There is potential to modulate VPS35 using viral vectors or genome editing techniques like CRISPR/Cas9, however, given VPS35's ubiquitous role in many homeostatic processes, strict dose control and regional specificity would be necessary to achieve a safe, therapeutic effect.[10] AAV2 vectors have demonstrated safety and efficacy in clinical trials and may be designed to introduce a high yield of non-mutant VPS35 to patients with neurodegenerative diseases.[11]
Speculative therapeutic applications for VPS35 include developing a biomarker assay that detects lower levels of VPS35 in PD or AD patients and identifying cis-regulatory elements within the VPS35 gene for microRNA therapy to reverse VPS35 deficiency.[5]
References[edit]
- ^ a b c d e f g h i j k l Reitz C (April 2015). "The role of the retromer complex in aging-related neurodegeneration: a molecular and genomic review". Molecular Genetics and Genomics. 290 (2): 413–27. doi:10.1007/s00438-014-0939-9. PMC 4363161. PMID 25332075.
- ^ a b c d e f g h i Reitz C (May 2018). "Retromer Dysfunction and Neurodegenerative Disease". Current Genomics. 19 (4): 279–288. doi:10.2174/1389202919666171024122809. PMC 5930449. PMID 29755290.
- ^ a b c d e f g h i j k l m n o Brodin L, Shupliakov O (2018). "Retromer in Synaptic Function and Pathology". Frontiers in Synaptic Neuroscience. 10: 37. doi:10.3389/fnsyn.2018.00037. PMC 6207580. PMID 30405388.
- ^ a b c d e Follett J, Bugarcic A, Collins BM, Teasdale RD (2017-07-01). "Retromer's Role in Endosomal Trafficking and Impaired Function in Neurodegenerative Diseases". Current Protein & Peptide Science. 18 (7): 687–701. doi:10.2174/1389203717666160311121246. PMID 26965691.
- ^ a b c d e f g h i j k l m n o Trousdale C, Kim K (November 2015). "Retromer: Structure, function, and roles in mammalian disease". European Journal of Cell Biology. 94 (11): 513–21. doi:10.1016/j.ejcb.2015.07.002. PMID 26220253.
- ^ a b c d e f g Vagnozzi AN, Li JG, Chiu J, Razmpour R, Warfield R, Ramirez SH, Praticò D (July 2019). "VPS35 regulates tau phosphorylation and neuropathology in tauopathy". Molecular Psychiatry. 26 (11): 6992–7005. doi:10.1038/s41380-019-0453-x. PMC 6949432. PMID 31289348.
- ^ a b c d e f g h i j k l m n Rahman AA, Morrison BE (March 2019). "Contributions of VPS35 Mutations to Parkinson's Disease". Neuroscience. 401: 1–10. doi:10.1016/j.neuroscience.2019.01.006. PMC 6422337. PMID 30660673.
- ^ a b c d e f g h i j k Vilariño-Güell C, Wider C, Ross OA, Dachsel JC, Kachergus JM, Lincoln SJ, et al. (July 2011). "VPS35 mutations in Parkinson disease". American Journal of Human Genetics. 89 (1): 162–7. doi:10.1016/j.ajhg.2011.06.001. PMC 3135796. PMID 21763482.
- ^ a b c d e f g h i j k l m n o p q r s t u v w x y z aa ab ac ad ae af ag Williams ET, Chen X, Moore DJ (2017-01-01). "VPS35, the Retromer Complex and Parkinson's Disease". Journal of Parkinson's Disease. 7 (2): 219–233. doi:10.3233/JPD-161020. PMC 5438477. PMID 28222538.
- ^ a b c d e f g h i j k l m n o p q r s t u v w x y z aa Cutillo G, Simon DK, Eleuteri S (November 2020). "VPS35 and the mitochondria: Connecting the dots in Parkinson's disease pathophysiology". Neurobiology of Disease. 145: 105056. doi:10.1016/j.nbd.2020.105056. PMID 32853677. S2CID 221277514.
- ^ a b c d e f g h i j k l m n o p q r s t u v w x y z aa ab ac ad Eleuteri S, Albanese A (2019-12-17). "VPS35-Based Approach: A Potential Innovative Treatment in Parkinson's Disease". Frontiers in Neurology. 10: 1272. doi:10.3389/fneur.2019.01272. PMC 6928206. PMID 31920908.
- ^ a b c GRCh38: Ensembl release 89: ENSG00000069329 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000031696 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ a b c d e f g h i j k l m n o Deng H, Gao K, Jankovic J (May 2013). "The VPS35 gene and Parkinson's disease". Movement Disorders. 28 (5): 569–75. doi:10.1002/mds.25430. PMID 23536430. S2CID 16641707.
- ^ a b c d e f g h i j k l m n o p q r s t u v w x y z aa Sassone J, Reale C, Dati G, Regoni M, Pellecchia MT, Garavaglia B (April 2020). "The Role of VPS35 in the Pathobiology of Parkinson's Disease". Cellular and Molecular Neurobiology. 41 (2): 199–227. doi:10.1007/s10571-020-00849-8. PMID 32323152. S2CID 216076582.
- ^ a b Vergés M (2016-01-01). Jeon KW (ed.). "Retromer in Polarized Protein Transport". International Review of Cell and Molecular Biology. 323. Academic Press: 129–79. doi:10.1016/bs.ircmb.2015.12.005. ISBN 9780128048085. PMID 26944621.
- ^ a b Ghosh R, Pattison JS (2018). "Macroautophagy and Chaperone-Mediated Autophagy in Heart Failure: The Known and the Unknown". Oxidative Medicine and Cellular Longevity. 2018: 8602041. doi:10.1155/2018/8602041. PMC 5822756. PMID 29576856.
- ^ a b c d e f g Bose A, Beal MF (October 2016). "Mitochondrial dysfunction in Parkinson's disease". Journal of Neurochemistry. 139 (S1): 216–231. doi:10.1111/jnc.13731. PMID 27546335. S2CID 32612919.
- ^ a b c d Weissbach A, Wittke C, Kasten M, Klein C (2019-01-01). Stamelou M, Höglinger GU (eds.). "'Atypical' Parkinson's disease - genetic". International Review of Neurobiology. 149. Academic Press: 207–235. doi:10.1016/bs.irn.2019.10.011. ISBN 9780128177303. PMID 31779813. S2CID 208357146.
- ^ a b Kim CY, Alcalay RN (April 2017). "Genetic Forms of Parkinson's Disease". Seminars in Neurology. 37 (2): 135–146. doi:10.1055/s-0037-1601567. PMID 28511254. S2CID 635105.
- ^ Lunati A, Lesage S, Brice A (November 2018). "The genetic landscape of Parkinson's disease" (PDF). Revue Neurologique. International SFN/SOMFA Meeting 2018. 174 (9): 628–643. doi:10.1016/j.neurol.2018.08.004. PMID 30245141.
- ^ a b Nguyen M, Wong YC, Ysselstein D, Severino A, Krainc D (February 2019). "Synaptic, Mitochondrial, and Lysosomal Dysfunction in Parkinson's Disease". Trends in Neurosciences. 42 (2): 140–149. doi:10.1016/j.tins.2018.11.001. PMC 6452863. PMID 30509690.
- ^ a b Abeysinghe AA, Deshapriya RD, Udawatte C (September 2020). "Alzheimer's disease; a review of the pathophysiological basis and therapeutic interventions". Life Sciences. 256: 117996. doi:10.1016/j.lfs.2020.117996. PMID 32585249. S2CID 220075087.
- ^ a b Boche D, Nicoll JA (December 2020). "Invited Review - Understanding cause and effect in Alzheimer's pathophysiology: Implications for clinical trials". Neuropathology and Applied Neurobiology. 46 (7): 623–640. doi:10.1111/nan.12642. PMID 32643143. S2CID 220429715.
- ^ Qureshi YH, Baez P, Reitz C (September 2020). "Endosomal Trafficking in Alzheimer's Disease, Parkinson's Disease, and Neuronal Ceroid Lipofuscinosis". Molecular and Cellular Biology. 40 (19). doi:10.1128/MCB.00262-20. PMC 7491951. PMID 32690545.
Further reading[edit]
- Maruyama K, Sugano S (January 1994). "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides". Gene. 138 (1–2): 171–4. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.
- Andersson B, Wentland MA, Ricafrente JY, Liu W, Gibbs RA (April 1996). "A "double adaptor" method for improved shotgun library construction". Analytical Biochemistry. 236 (1): 107–13. doi:10.1006/abio.1996.0138. PMID 8619474.
- Bonaldo MF, Lennon G, Soares MB (September 1996). "Normalization and subtraction: two approaches to facilitate gene discovery". Genome Research. 6 (9): 791–806. doi:10.1101/gr.6.9.791. PMID 8889548.
- Yu W, Andersson B, Worley KC, Muzny DM, Ding Y, Liu W, et al. (April 1997). "Large-scale concatenation cDNA sequencing". Genome Research. 7 (4): 353–8. doi:10.1101/gr.7.4.353. PMC 139146. PMID 9110174.
- Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, Suyama A, Sugano S (October 1997). "Construction and characterization of a full length-enriched and a 5'-end-enriched cDNA library". Gene. 200 (1–2): 149–56. doi:10.1016/S0378-1119(97)00411-3. PMID 9373149.
- Edgar AJ, Polak JM (November 2000). "Human homologues of yeast vacuolar protein sorting 29 and 35". Biochemical and Biophysical Research Communications. 277 (3): 622–30. doi:10.1006/bbrc.2000.3727. PMID 11062004.
- Hartley JL, Temple GF, Brasch MA (November 2000). "DNA cloning using in vitro site-specific recombination". Genome Research. 10 (11): 1788–95. doi:10.1101/gr.143000. PMC 310948. PMID 11076863.
- Haft CR, de la Luz Sierra M, Bafford R, Lesniak MA, Barr VA, Taylor SI (December 2000). "Human orthologs of yeast vacuolar protein sorting proteins Vps26, 29, and 35: assembly into multimeric complexes". Molecular Biology of the Cell. 11 (12): 4105–16. doi:10.1091/mbc.11.12.4105. PMC 15060. PMID 11102511.
- Vergés M, Luton F, Gruber C, Tiemann F, Reinders LG, Huang L, et al. (August 2004). "The mammalian retromer regulates transcytosis of the polymeric immunoglobulin receptor". Nature Cell Biology. 6 (8): 763–9. doi:10.1038/ncb1153. PMID 15247922. S2CID 22296469.
- Mingot JM, Bohnsack MT, Jäkle U, Görlich D (August 2004). "Exportin 7 defines a novel general nuclear export pathway". The EMBO Journal. 23 (16): 3227–36. doi:10.1038/sj.emboj.7600338. PMC 514512. PMID 15282546.