| Myosin Light-Chain Phosphatase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Structure of complex between PP1 and a portion of MYPT1, generated from 1s70[1] | |||||||||
| Identifiers | |||||||||
| EC no. | 3.1.3.53 | ||||||||
| CAS no. | 86417-96-1 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Myosin light-chain phosphatase, also called myosin phosphatase (EC 3.1.3.53; systematic name [myosin-light-chain]-phosphate phosphohydrolase), is an enzyme (specifically a serine/threonine-specific protein phosphatase) that dephosphorylates the regulatory light chain of myosin II:
- [myosin light-chain] phosphate + H2O = [myosin light-chain] + phosphate
This dephosphorylation reaction occurs in smooth muscle tissue and initiates the relaxation process of the muscle cells. Thus, myosin phosphatase undoes the muscle contraction process initiated by myosin light-chain kinase. The enzyme is composed of three subunits: the catalytic region (protein phosphatase 1, or PP1), the myosin binding subunit (MYPT1), and a third subunit (M20) of unknown function. The catalytic region uses two manganese ions as catalysts to dephosphorylate the light-chains on myosin, which causes a conformational change in the myosin and relaxes the muscle. The enzyme is highly conserved[1] and is found in all organisms’ smooth muscle tissue. While it is known that myosin phosphatase is regulated by rho-associated protein kinases, there is current debate about whether other molecules, such as arachidonic acid and cAMP, also regulate the enzyme.[2]
Function[edit]
Smooth muscle tissue is mostly made of actin and myosin,[3] two proteins that interact together to produce muscle contraction and relaxation. Myosin II, also known as conventional myosin, has two heavy chains that consist of the head and tail domains and four light chains (two per head) that bind to the heavy chains in the “neck” region. When the muscle needs to contract, calcium ions flow into the cytosol from the sarcoplasmic reticulum, where they activate calmodulin, which in turn activates myosin light-chain kinase (MLC kinase). MLC kinase phosphorylates the myosin light chain (MLC20) at the Ser-19 residue. This phosphorylation causes a conformational change in the myosin, activating crossbridge cycling and causing the muscle to contract. Because myosin undergoes a conformational change, the muscle will stay contracted even if calcium and activated MLC kinase concentrations are brought to normal levels. The conformational change must be undone to relax the muscle.[4]
When myosin phosphatase binds to myosin, it removes the phosphate group. Without the group, the myosin reverts to its original conformation, in which it cannot interact with the actin and hold the muscle tense, so the muscle relaxes. The muscle will remain in this relaxed position until myosin is phosphorylated by MLC kinase and undergoes a conformational change.
Structure[edit]
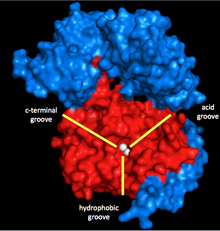
Myosin phosphatase is made of three subunits. The catalytic subunit, PP1, is one of the more important Ser/Thr phosphatases in eukaryotic cells, as it plays a role in glycogen metabolism, intracellular transport, protein synthesis, and cell division as well as smooth muscle contraction.[5] Because it is so important to basic cellular functions, and because there are far fewer protein phosphatases than kinases in cells,[6] PP1’s structure and function is highly conserved (though the specific isoform used in myosin phosphatase is the δ isoform, PP1δ).[7] PP1 works by using two manganese ions as catalysts for the dephosphorylation (see below).
Surrounding these ions is a Y-shaped cleft with three grooves: a hydrophobic, an acidic, and a C-terminal groove. When PP1 is not bonded to any other subunit, it is not particularly specific. However, when it bonds to the second subunit of myosin phosphatase, MYPT1 (MW ~130 kDa), this catalytic cleft changes configuration. This results in a dramatic increase in myosin specificity.[1] Thus, it is clear that MYPT1 has great regulatory power over PP1 and myosin phosphatase, even without the presence of other activators or inhibitors.
The third subunit, M20 (not to be confused with MLC20, the critical regulatory subunit of myosin), is the smallest and most mysterious subunit. Currently little is known about M20, except that it is not necessary for catalysis, as removing the subunit does not affect turnover or selectivity.[1] While some believe it could have regulatory function, nothing has been determined yet.[2]
Mechanism[edit]
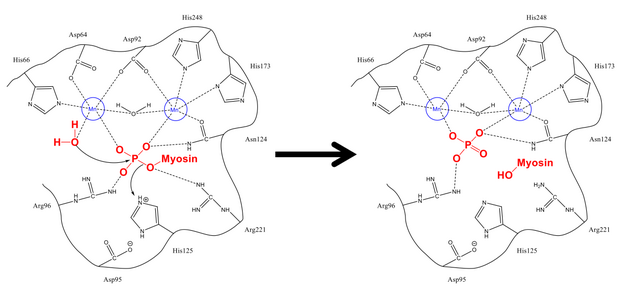
The mechanism of removing the phosphate from Ser-19 is very similar to other dephosphorylation reactions in the cell, such as the activation of glycogen synthase. Myosin's regulatory subunit MLC20 binds to both the hydrophobic and acid grooves of PP1 and MYPT1, the regulatory site on myosin phosphatase.[1][8] Once in the proper configuration, both the phyosphorylated serine and a free water molecule are stabilized by the hydrogen-bonding residues in the active site, as well as the positively charged ions (which interact strongly with the negative phosphate group). His-125 (on myosin phosphatase) donates a proton to Ser-19 MLC20), and the water molecule attacks the phosphorus atom. After shuffling protons to stabilize (which happens rapidly compared to the attack on phosphorus), the phosphate and alcohol are formed, and both leave the active site.
Regulation and Human Health[edit]
The regulatory pathways of MLC kinase have been well-established, but until the late 1980s, it was assumed that myosin phosphatase was not regulated, and contraction/relaxation was entirely dependent on MLC kinase activity.[2] However, since the 1980s, the inhibiting effect of rho-associated protein kinase has been discovered and thoroughly investigated.[11] RhoA GTP activates Rho-kinase, which phosphorylates the MYPT1 at two major inhibitory sites, Thr-696 and Thr-866.[12][13] This fully demonstrates the value of the MYPT1, not only to increase reaction rate and specificity, but also to greatly slow down the reaction. However, when telokin is added, it effectively undoes the effect of Rho-kinase, even though it does not dephosphorylate MYPT1.[12]
One other proposed regulatory strategy involves arachidonic acid. When arachidonic acid is added to tensed muscle tissue, the acid decreases the rate of dephosphorylation (and thus relaxation) of myosin. However, it is unclear how arachidonic acid functions as an inhibitor.[4] Two competing theories are that either arachidonic acid acts as a co-messenger in the rho-kinase cascade mentioned above, or that it binds to the c-terminal of MYPT1.[4]
When the regulatory systems of myosin phosphatase begin to fail, there can be major health consequences. Since smooth muscle is found in the respiratory, circulatory, and reproductive systems of humans (as well as other places), if the smooth muscle can no longer relax because of faulty regulation, then a wide number of problems ranging from asthma, hypertension, and erectile dysfunction can result.[4][14]
See also[edit]
References[edit]
- ^ a b c d e Terrak, Mohammed; Kerff, Frederic; et al. (June 17, 2004). "Structural Basis of Protein Phosphatase 1 Regulation". Nature. 429 (6993): 780–4. Bibcode:2004Natur.429..780T. doi:10.1038/nature02582. PMID 15164081.
- ^ a b c Hartshorne, DJ; Ito, M (May 1998). "Myosin Light Chain Phosphatase: Subunit Composition, Interactions and Regulation". J Muscle Res Cell Motil. 19 (4): 325–41. doi:10.1023/A:1005385302064. PMID 9635276. S2CID 27448238.
- ^ Page 174 in: The vascular smooth muscle cell: molecular and biological responses to the extracellular matrix. Authors: Stephen M. Schwartz, Robert P. Mecham. Editors: Stephen M. Schwartz, Robert P. Mecham. Contributors: Stephen M. Schwartz, Robert P. Mecham. Publisher: Academic Press, 1995. ISBN 0-12-632310-0, ISBN 978-0-12-632310-8
- ^ a b c d Webb, R. Clinton (November 2003). "Smooth Muscle Contraction and Relaxation". Advances in Physiology Education. 27 (4): 201–6. doi:10.1152/advan.00025.2003. PMID 14627615.
- ^ Hurley, Thomas; Yang, Jie; et al. (July 18, 2007). "Structural Basis for Regulation of Protein Phosphatase 1 by Inhibitor-2". J. Biol. Chem. 282 (39): 28874–83. doi:10.1074/jbc.m703472200. PMID 17636256.
- ^ Cohen, Patricia T. W. (January 15, 2002). "Protein Phosphatase 1-Targeted in Many Directions". J Cell Sci. 115 (2): 780–4. doi:10.1242/jcs.115.2.241. PMID 11839776.
- ^ Fujioka, M; Takahashi, N (April 1, 1998). "A New Isoform of Human Myosin Phosphatase Targeting/Regulatory Subunit (MYPT2): cDNA Cloning, Tissue Expression, and Chromosomal Mapping". Genomics. 49 (1): 325–41. doi:10.1006/geno.1998.5222. PMID 9570949.
- ^ Gomperts, Bastein D. (August 19, 2009). Signal Transduction: 2nd Edition. London: Academic Press. ISBN 978-0123694416.
- ^ Shi, Yigong (October 30, 2009). "Serine/Threonine Phosphatases: Mechanism through Structure". Cell. 139 (3): 468–84. doi:10.1016/j.cell.2009.10.006. PMID 19879837. S2CID 13903804.
- ^ Lee, Ernest Y.C.; Zhang, Lifang; et al. (March 15, 1999). "Phosphorylase Phosphatase: New Horizons for an Old Enzyme". Frontiers in Bioscience. 4 (1–3): 270–85. doi:10.2741/lee. PMID 10077543. Retrieved March 9, 2015.
- ^ Wang, Yuepeng; Riddick, Nadeen; et al. (February 27, 2009). "ROCK Isoform Regulation of Myosin Phosphatase and Contractility in Vascular Smooth Muscle Cells". Circ. Res. 104 (4): 531–40. doi:10.1161/circresaha.108.188524. PMC 2649695. PMID 19131646.
- ^ a b Khromov, ES; Momotani, K.; et al. (April 27, 2012). "Molecular Mechanism of Telokin-Mediated Disinhibition of Myosin Light Chain Phosphatase and cAMP/cGMP-Induced Relaxation of Gastrointestinal Smooth Muscle". J Biol Chem. 287 (25): 20975–85. doi:10.1074/jbc.m112.341479. PMC 3375521. PMID 22544752.
- ^ Somlyo, Andrew P.; Somlyo, Avril V. (November 10, 1999). "Signal Transduction by G-Proteins, Rho-Kinase and Protein Phosphatase to Smooth Muscle and Non-Muscle Myosin II". Journal of Physiology. 522 (2): 177–85. doi:10.1111/j.1469-7793.2000.t01-2-00177.x. PMC 2269761. PMID 10639096.
- ^ Aguilar, Hector; Mitchell, B.F. (May 7, 2010). "Physiological Pathways and Molecular Mechanisms Regulating Uterine Contractility". Human Reproduction Update. 16 (6): 725–44. doi:10.1093/humupd/dmq016. PMID 20551073.
Further reading[edit]
- Pato MD, Adelstein RS (1983). "Purification and characterization of a multisubunit phosphatase from turkey gizzard smooth muscle. The effect of calmodulin binding to myosin light chain kinase on dephosphorylation". J. Biol. Chem. 258 (11): 7047–54. doi:10.1016/S0021-9258(18)32330-5. PMID 6304072.
- Kimura K; et al. (1996). "Regulation of Myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase)". Science. 273 (5272): 245–248. Bibcode:1996Sci...273..245K. doi:10.1126/science.273.5272.245. PMID 8662509. S2CID 37249779.