| |
| Clinical data | |
|---|---|
| Trade names | Oxigon[2] |
| Other names | Conjugate acid: • 1H-Indole-3-propanoic acid • Indole-3-propionic acid Conjugate base: • Indole-3-propionate |
| ATC code |
|
| Legal status | |
| Legal status |
|
| Identifiers | |
| |
| CAS Number | |
| PubChem CID | |
| IUPHAR/BPS | |
| ChemSpider | |
| UNII | |
| ChEBI | |
| CompTox Dashboard (EPA) | |
| ECHA InfoCard | 100.011.455 |
| Chemical and physical data | |
| Formula | C11H11NO2 |
| Molar mass | 189.214 g·mol−1 |
| 3D model (JSmol) | |
| Melting point | 134 to 135 °C (273 to 275 °F) [3] |
| |
| |
| (verify) | |
3-Indolepropionic acid (IPA), or indole-3-propionic acid, has been studied for its therapeutic therapeutic value in the treatment of Alzheimer's disease. As of 2022[4] IPA shows potential in the treatment of this disease, though the therapeutic effect of IPA depends on dose and time of therapy initiation.
Though promising in some historical clinical trials, IPA is not clinically listed as a useful therapeutic in managing Alzheimer's as of 2023.[5]
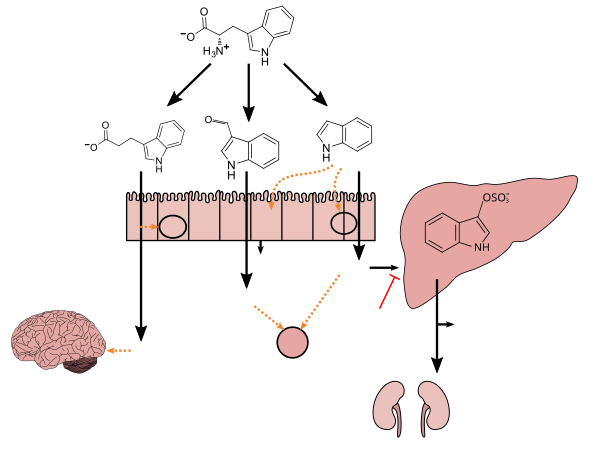
This compound endogenously produced by human microbiota and has only been detected in vivo when the species Clostridium sporogenes is present in the gastrointestinal tract.[6][7][8] As of April 2016[update], C. sporogenes, which uses tryptophan to synthesize IPA, is the only species of bacteria known to synthesize IPA in vivo at levels which are subsequently detectable in the blood plasma of the host.[6][7][8][9]
IPA is an even more potent scavenger of hydroxyl radicals than melatonin, the most potent scavenger of hydroxyl radicals that is synthesized by human enzymes.[3][9] Similar to melatonin but unlike other antioxidants, it scavenges radicals without subsequently generating reactive and pro-oxidant intermediate compounds.[3][9][10] In 2017, elevated concentrations of IPA in human blood plasma were found to be correlated with a lower risk of type 2 diabetes and higher consumption of fiber-rich foods.[3][11][12]
Biosynthesis in humans and cellular effects[edit]
Tryptophan metabolism by human gastrointestinal microbiota ()
|
Metabolism[edit]
IPA can be converted in the liver or kidneys to 3-indoleacrylic acid, which is subsequently conjugated with glycine, forming indolylacryloyl glycine.[13]
History[edit]
The neuroprotective, antioxidant, and anti-amyloid properties of IPA were first reported in 1999.[9][14][15][16]
See also[edit]
References[edit]
- ^ Galligan JJ (February 2018). "Beneficial actions of microbiota-derived tryptophan metabolites". Neurogastroenterology and Motility. 30 (2): e13283. doi:10.1111/nmo.13283. PMID 29341448. S2CID 39904059.
- ^ Bendheim PE, Poeggeler B, Neria E, Ziv V, Pappolla MA, Chain DG (October 2002). "Development of indole-3-propionic acid (OXIGON) for Alzheimer's disease". Journal of Molecular Neuroscience. 19 (1–2): 213–217. doi:10.1007/s12031-002-0036-0. PMID 12212784. S2CID 31107810.
The accumulation of amyloid-beta and concomitant oxidative stress are major pathogenic events in Alzheimer's disease. Indole-3-propionic acid (IPA, OXIGON) is a potent anti-oxidant devoid of pro-oxidant activity. IPA has been demonstrated to be an inhibitor of beta-amyloid fibril formation and to be a potent neuroprotectant against a variety of oxidotoxins. This review will summarize the known properties of IPA and outline the rationale behind its selection as a potential disease-modifying therapy for Alzheimer's disease.
- ^ a b c d e f "3-Indolepropionic acid". Human Metabolome Database. University of Alberta. Retrieved 12 June 2018.
- ^ Jiang H, Chen C, Gao J (December 2022). "Extensive Summary of the Important Roles of Indole Propionic Acid, a Gut Microbial Metabolite in Host Health and Disease". Nutrients. 15 (1): 151. doi:10.3390/nu15010151. PMC 9824871. PMID 36615808.
- ^ "How Alzheimer's drugs help manage symptoms". Mayo Clinic. Retrieved 3 November 2023.
- ^ a b c Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G (March 2009). "Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites". Proc. Natl. Acad. Sci. U.S.A. 106 (10): 3698–3703. Bibcode:2009PNAS..106.3698W. doi:10.1073/pnas.0812874106. PMC 2656143. PMID 19234110.
Production of IPA was shown to be completely dependent on the presence of gut microflora and could be established by colonization with the bacterium Clostridium sporogenes. ... Conversely, a different set of enteric bacteria has been implicated in the metabolic transformation of indole to indole-3-propionic acid (IPA) (27). IPA, also identified only in the plasma of conv mice, has been shown to be a powerful antioxidant (28) ... Although the presence of IPA in mammals has long been ascribed in the literature to bacterial metabolic processes, this conclusion was based on either the production of IPA in ex vivo cultures of individual bacterial species (31) or observed decreases in IPA levels in animals after administration of antibiotics (32). In our own survey of IPA production by representative members of the intestinal flora, only Clostridium sporogenes was found to produce IPA in culture (Table S2). Based on these results, individual GF mice were intentionally colonized with C. sporogenes strain ATCC 15579, and blood samples were taken at several intervals after colonization. IPA was undetectable in the samples taken shortly after introduction of the microbes, and was first observed in the serum 5 days after colonization, reaching plateau values comparable with conv mice by day 10. These colonization studies demonstrate that the introduction of enteric bacteria capable of IPA production in vivo into the gastrointestinal tract is sufficient to introduce IPA into the bloodstream of the host. Also, other GF animals were injected i.p. with either IPA (at 10, 20, or 40 mg/kg) or sterile PBS vehicle, and their serum concentrations of IPA were measured over time. As seen in Table S3, the high serum levels of IPA observed 1 h after injection decreased more than 90% within 5 h, showing that IPA is rapidly cleared from the blood, and that its presence in the serum of conv animals must result from continuous production from 1 or more bacterial species associated with the mammalian gut.
IPA metabolism diagram - ^ a b c d e f g h i j k Zhang LS, Davies SS (April 2016). "Microbial metabolism of dietary components to bioactive metabolites: opportunities for new therapeutic interventions". Genome Med. 8 (1): 46. doi:10.1186/s13073-016-0296-x. PMC 4840492. PMID 27102537.
Lactobacillus spp. convert tryptophan to indole-3-aldehyde (I3A) through unidentified enzymes [125]. Clostridium sporogenes convert tryptophan to IPA [6], likely via a tryptophan deaminase. ... IPA also potently scavenges hydroxyl radicals
Table 2: Microbial metabolites: their synthesis, mechanisms of action, and effects on health and disease
Figure 1: Molecular mechanisms of action of indole and its metabolites on host physiology and disease - ^ a b Attwood G, Li D, Pacheco D, Tavendale M (June 2006). "Production of indolic compounds by rumen bacteria isolated from grazing ruminants". Journal of Applied Microbiology. 100 (6): 1261–1271. doi:10.1111/j.1365-2672.2006.02896.x. PMID 16696673. S2CID 35673610.
- ^ a b c d e Chyan YJ, Poeggeler B, Omar RA, Chain DG, Frangione B, Ghiso J, Pappolla MA (July 1999). "Potent neuroprotective properties against the Alzheimer beta-amyloid by an endogenous melatonin-related indole structure, indole-3-propionic acid". J. Biol. Chem. 274 (31): 21937–21942. doi:10.1074/jbc.274.31.21937. PMID 10419516. S2CID 6630247.
[Indole-3-propionic acid (IPA)] has previously been identified in the plasma and cerebrospinal fluid of humans, but its functions are not known. ... In kinetic competition experiments using free radical-trapping agents, the capacity of IPA to scavenge hydroxyl radicals exceeded that of melatonin, an indoleamine considered to be the most potent naturally occurring scavenger of free radicals. In contrast with other antioxidants, IPA was not converted to reactive intermediates with pro-oxidant activity.
- ^ Reiter RJ, Guerrero JM, Garcia JJ, Acuña-Castroviejo D (November 1998). "Reactive oxygen intermediates, molecular damage, and aging. Relation to melatonin". Annals of the New York Academy of Sciences. 854 (1): 410–424. Bibcode:1998NYASA.854..410R. doi:10.1111/j.1749-6632.1998.tb09920.x. PMID 9928448. S2CID 29333394.
- ^ de Mello VD, Paananen J, Lindström J, Lankinen MA, Shi L, Kuusisto J, et al. (April 2017). "Indolepropionic acid and novel lipid metabolites are associated with a lower risk of type 2 diabetes in the Finnish Diabetes Prevention Study". Scientific Reports. 7: 46337. Bibcode:2017NatSR...746337D. doi:10.1038/srep46337. PMC 5387722. PMID 28397877.
- ^ Tuomainen M, Lindström J, Lehtonen M, Auriola S, Pihlajamäki J, Peltonen M, et al. (May 2018). "Associations of serum indolepropionic acid, a gut microbiota metabolite, with type 2 diabetes and low-grade inflammation in high-risk individuals". Nutrition & Diabetes. 8 (1): 35. doi:10.1038/s41387-018-0046-9. PMC 5968030. PMID 29795366.
- ^ Keszthelyi D, Troost FJ, Masclee AA (December 2009). "Understanding the role of tryptophan and serotonin metabolism in gastrointestinal function". Neurogastroenterology and Motility. 21 (12): 1239–1249. doi:10.1111/j.1365-2982.2009.01370.x. PMID 19650771. S2CID 23568813.
Indolylpropionic acid can be further converted in the liver or kidney into indolyl acrylic acid (IAcrA) and conjugated with glycine to produce indolylacryloyl glycine (IAcrGly). ... Also, indolyl propionic acid has been shown to be a powerful antioxidant, and is currently being investigated as a possible treatment for Alzheimer's disease.40
- ^ Poeggeler B, Sambamurti K, Siedlak SL, Perry G, Smith MA, Pappolla MA (April 2010). "A novel endogenous indole protects rodent mitochondria and extends rotifer lifespan". PLOS ONE. 5 (4): e10206. Bibcode:2010PLoSO...510206P. doi:10.1371/journal.pone.0010206. PMC 2858081. PMID 20421998.
- ^ Karbownik M, Reiter RJ, Garcia JJ, Cabrera J, Burkhardt S, Osuna C, Lewiński A (2001). "Indole-3-propionic acid, a melatonin-related molecule, protects hepatic microsomal membranes from iron-induced oxidative damage: relevance to cancer reduction". Journal of Cellular Biochemistry. 81 (3): 507–513. doi:10.1002/1097-4644(20010601)81:3<507::AID-JCB1064>3.0.CO;2-M. PMID 11255233. S2CID 27462000.
- ^ Reiter RJ, Tan DX, Osuna C, Gitto E (2000). "Actions of melatonin in the reduction of oxidative stress. A review". Journal of Biomedical Science. 7 (6): 444–458. doi:10.1007/bf02253360. PMID 11060493.