| GRM3 | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | GRM3, GLUR3, GPRC1C, MGLUR3, mGlu3, glutamate metabotropic receptor 3 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 601115 MGI: 1351340 HomoloGene: 651 GeneCards: GRM3 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Metabotropic glutamate receptor 3 (mGluR3) is an inhibitory Gi/G0-coupled G-protein coupled receptor (GPCR)[4] generally localized to presynaptic sites of neurons in classical circuits.[5] However, in higher cortical circuits in primates, mGluR3 are localized post-synaptically, where they strengthen rather than weaken synaptic connectivity.[6] In humans, mGluR3 is encoded by the GRM3 gene.[7][8] Deficits in mGluR3 signaling have been linked to impaired cognition in humans,[9] and to increased risk of schizophrenia,[10] consistent with their expanding role in cortical evolution.
Structure[edit]
In humans, mGluR3 is encoded by the GRM3 gene on chromosome 7. At least five protein-coding isoforms are predicted based on genomic information. The mGluR3 protein is a seven-pass transmembrane protein.
Function[edit]
L-glutamate is the major excitatory neurotransmitter in the central nervous system and activates both ionotropic and metabotropic glutamate receptors. Glutamatergic neurotransmission is involved in most aspects of normal brain function and can be perturbed in many neuropathologic conditions. The metabotropic glutamate receptors are a family of G protein-coupled receptors, that have been divided into 3 groups on the basis of sequence homology, putative signal transduction mechanisms, and pharmacologic properties. Group I includes GRM1 and GRM5 and these receptors have been shown to activate phospholipase C. Group II includes GRM2 and GRM3 while Group III includes GRM4, GRM6, GRM7 and GRM8. Group II and III receptors are linked to the inhibition of the cyclic AMP cascade but differ in their agonist selectivities.[8]
Clinical significance[edit]
The mGluR3 receptor encoded by the GRM3 gene has been found to be associated with a range of psychiatric disorders, including bipolar affective disorder[11] as well as schizophrenia.[12][10]
A mutation in the Kozak sequence in the 1st exon of the GRM3 gene was shown to change translation and transcription of cloned GRM3 gene constructs and was significantly associated with bipolar disorder with an odds ratio of 4.4.[11] Subsequently, a marker in GRM3 was implicated in a large genome-wide association study of schizophrenia with statistical significance of p<10−9.[13] A follow-up study of the Kozak sequence variant showed that it was associated with increased risk of bipolar disorder, schizophrenia and alcoholism.[14] The mGluR3 receptor encoded by GRM3 is targetable by several drugs that have been used in previous trials of schizophrenia and anxiety disorder. The agonist, antagonist and allosteric modulator drugs of mGluR3 can now be explored as new treatments for mental illness.[11] Other scientific evidence has been published which shows that the well established anti-manic drug lithium carbonate also changes GRM3 gene expression in the mouse brain after treatment with lithium carbonate.[15]
Ligands[edit]
mGluR3 modulators that are significantly selective over the isoform mGluR2 are known since 2013.
Agonists[edit]
- with a bicyclo[3.1.0]hexane skeleton
- (R)-2-amino-4-(4-hydroxy[1,2,5]thiadiazol-3-yl)butyric acid[22]
Antagonists[edit]
- CECXG – 38x selectivity for mGlu3 over mGlu2
- LY-341,495 and its 1-fluoro analog:[23] potent orthosteric antagonists
- MGS-0039,[24] HYDIA[25] (both with bicyclo[3.1.0]hexane skeleton)
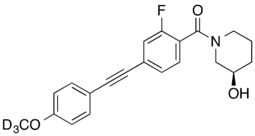
Allosteric modulators[edit]
- D3-ML337: selective NAM, IC50 = 450 nM for mGluR3, IC50 >30μM for mGluR2[26]
- MNI-137:[27] inhibitor
- VU-0650786: NAM[28]
- compound 7p:[29] non-competitive antagonist (presumably allosteric inhibitor)
- LY 2389575: negative allosteric modulator.[30]
Interactions[edit]
Metabotropic glutamate receptor 3 has been shown to interact with:
See also[edit]
References[edit]
- ^ a b c GRCh38: Ensembl release 89: ENSG00000198822 - Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Ambrosini A, Bresciani L, Fracchia S, Brunello N, Racagni G (May 1995). "Metabotropic glutamate receptors negatively coupled to adenylate cyclase inhibit N-methyl-D-aspartate receptor activity and prevent neurotoxicity in mesencephalic neurons in vitro" (abstract). Molecular Pharmacology. 47 (5): 1057–64. PMID 7746273.
- ^ Shigemoto R, Kinoshita A, Wada E, Nomura S, Ohishi H, Takada M, et al. (October 1997). "Differential presynaptic localization of metabotropic glutamate receptor subtypes in the rat hippocampus". The Journal of Neuroscience. 17 (19): 7503–22. doi:10.1523/JNEUROSCI.17-19-07503.1997. PMC 6573434. PMID 9295396.
- ^ Jin LE, Wang M, Galvin VC, Lightbourne TC, Conn PJ, Arnsten AF, Paspalas CD (March 2018). "mGluR2 versus mGluR3 Metabotropic Glutamate Receptors in Primate Dorsolateral Prefrontal Cortex: Postsynaptic mGluR3 Strengthen Working Memory Networks". Cerebral Cortex. 28 (3): 974–987. doi:10.1093/cercor/bhx005. PMC 5974790. PMID 28108498.
- ^ Scherer SW, Duvoisin RM, Kuhn R, Heng HH, Belloni E, Tsui LC (January 1996). "Localization of two metabotropic glutamate receptor genes, GRM3 and GRM8, to human chromosome 7q". Genomics. 31 (2): 230–3. doi:10.1006/geno.1996.0036. PMID 8824806.
- ^ a b "Entrez Gene: GRM3 glutamate receptor, metabotropic 3".
- ^ Zink CF, Barker PB, Sawa A, Weinberger DR, Wang M, Quillian H, Ulrich WS, Chen Q, Jaffe AE, Kleinman JE, Hyde TM, Prettyman GE, Giegerich M, Carta K, van Ginkel M, Bigos KL (December 2020). "Association of Missense Mutation in FOLH1 With Decreased NAAG Levels and Impaired Working Memory Circuitry and Cognition". The American Journal of Psychiatry. 177 (12): 1129–1139. doi:10.1176/appi.ajp.2020.19111152. PMID 33256444. S2CID 227243375.
- ^ a b Saini SM, Mancuso SG, Mostaid MS, Liu C, Pantelis C, Everall IP, Bousman CA (August 2017). "Meta-analysis supports GWAS-implicated link between GRM3 and schizophrenia risk". Translational Psychiatry. 7 (8): e1196. doi:10.1038/tp.2017.172. PMC 5611739. PMID 28786982.
- ^ a b c Kandaswamy R, McQuillin A, Sharp SI, Fiorentino A, Anjorin A, Blizard RA, et al. (June 2013). "Genetic association, mutation screening, and functional analysis of a Kozak sequence variant in the metabotropic glutamate receptor 3 gene in bipolar disorder". JAMA Psychiatry. 70 (6): 591–8. doi:10.1001/jamapsychiatry.2013.38. PMID 23575746.
- ^ Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, et al. (February 2014). "De novo mutations in schizophrenia implicate synaptic networks". Nature. 506 (7487): 179–84. Bibcode:2014Natur.506..179F. doi:10.1038/nature12929. PMC 4237002. PMID 24463507.
- ^ Ripke S, Neale BM, Corvin A, Walters JT, Farh KH, Holmans PA, et al. (Schizophrenia Working Group of the Psychiatric Genomics Consortium) (July 2014). "Biological insights from 108 schizophrenia-associated genetic loci". Nature. 511 (7510): 421–7. Bibcode:2014Natur.511..421S. doi:10.1038/nature13595. PMC 4112379. PMID 25056061.
- ^ O'Brien NL, Way MJ, Kandaswamy R, Fiorentino A, Sharp SI, Quadri G, et al. (December 2014). "The functional GRM3 Kozak sequence variant rs148754219 affects the risk of schizophrenia and alcohol dependence as well as bipolar disorder". Psychiatric Genetics. 24 (6): 277–8. doi:10.1097/YPG.0000000000000050. PMC 4272221. PMID 25046171.
- ^ McQuillin A, Rizig M, Gurling HM (August 2007). "A microarray gene expression study of the molecular pharmacology of lithium carbonate on mouse brain mRNA to understand the neurobiology of mood stabilization and treatment of bipolar affective disorder". Pharmacogenetics and Genomics. 17 (8): 605–17. doi:10.1097/FPC.0b013e328011b5b2. PMID 17622937. S2CID 31180768.
- ^ Nakazato A, Kumagai T, Sakagami K, Yoshikawa R, Suzuki Y, Chaki S, et al. (December 2000). "Synthesis, SARs, and pharmacological characterization of 2-amino-3 or 6-fluorobicyclo[3.1.0]hexane-2,6-dicarboxylic acid derivatives as potent, selective, and orally active group II metabotropic glutamate receptor agonists". Journal of Medicinal Chemistry. 43 (25): 4893–909. doi:10.1021/jm000346k. PMID 11123999.
- ^ Monn JA, Massey SM, Valli MJ, Henry SS, Stephenson GA, Bures M, et al. (January 2007). "Synthesis and metabotropic glutamate receptor activity of S-oxidized variants of (-)-4-amino-2-thiabicyclo-[3.1.0]hexane-4,6-dicarboxylate: identification of potent, selective, and orally bioavailable agonists for mGlu2/3 receptors". Journal of Medicinal Chemistry. 50 (2): 233–40. doi:10.1021/jm060917u. PMID 17228865.
- ^ Monn JA, Valli MJ, Massey SM, Hansen MM, Kress TJ, Wepsiec JP, et al. (March 1999). "Synthesis, pharmacological characterization, and molecular modeling of heterobicyclic amino acids related to (+)-2-aminobicyclo[3.1.0] hexane-2,6-dicarboxylic acid (LY354740): identification of two new potent, selective, and systemically active agonists for group II metabotropic glutamate receptors". Journal of Medicinal Chemistry. 42 (6): 1027–40. doi:10.1021/jm980616n. PMID 10090786.
- ^ Monn JA, Valli MJ, Massey SM, Wright RA, Salhoff CR, Johnson BG, et al. (February 1997). "Design, synthesis, and pharmacological characterization of (+)-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylic acid (LY354740): a potent, selective, and orally active group 2 metabotropic glutamate receptor agonist possessing anticonvulsant and anxiolytic properties". Journal of Medicinal Chemistry. 40 (4): 528–37. doi:10.1021/jm9606756. PMID 9046344.
- ^ Dominguez C, Prieto L, Valli MJ, Massey SM, Bures M, Wright RA, et al. (May 2005). "Methyl substitution of 2-aminobicyclo[3.1.0]hexane 2,6-dicarboxylate (LY354740) determines functional activity at metabotropic glutamate receptors: identification of a subtype selective mGlu2 receptor agonist". Journal of Medicinal Chemistry. 48 (10): 3605–12. doi:10.1021/jm040222y. PMID 15887967.
- ^ Monn JA, Henry SS, Massey SM, Clawson DK, Chen Q, Diseroad BA, et al. (March 2018). "Synthesis and Pharmacological Characterization of C4β-Amide-Substituted 2-Aminobicyclo[3.1.0]hexane-2,6-dicarboxylates. Identification of (1 S,2 S,4 S,5 R,6 S)-2-Amino-4-[(3-methoxybenzoyl)amino]bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid (LY2794193), a Highly Potent and Selective mGlu3 Receptor Agonist". Journal of Medicinal Chemistry. 61 (6): 2303–2328. doi:10.1021/acs.jmedchem.7b01481. PMID 29350927.
- ^ Clausen RP, Bräuner-Osborne H, Greenwood JR, Hermit MB, Stensbøl TB, Nielsen B, Krogsgaard-Larsen P (September 2002). "Selective agonists at group II metabotropic glutamate receptors: synthesis, stereochemistry, and molecular pharmacology of (S)- and (R)-2-amino-4-(4-hydroxy[1,2,5]thiadiazol-3-yl)butyric acid". Journal of Medicinal Chemistry. 45 (19): 4240–5. doi:10.1021/jm020122x. PMID 12213064.
- ^ Sakagami K, Yasuhara A, Chaki S, Yoshikawa R, Kawakita Y, Saito A, et al. (April 2008). "Synthesis, in vitro pharmacology, and pharmacokinetic profiles of 2-[1-amino-1-carboxy-2-(9H-xanthen-9-yl)-ethyl]-1-fluorocyclopropanecarboxylic acid and its 6-heptyl ester, a potent mGluR2 antagonist". Bioorganic & Medicinal Chemistry. 16 (8): 4359–66. doi:10.1016/j.bmc.2008.02.066. PMID 18348906.
- ^ a) Nakazato A, Sakagami K, Yasuhara A, Ohta H, Yoshikawa R, Itoh M, et al. (August 2004). "Synthesis, in vitro pharmacology, structure-activity relationships, and pharmacokinetics of 3-alkoxy-2-amino-6-fluorobicyclo[3.1.0]hexane-2,6-dicarboxylic acid derivatives as potent and selective group II metabotropic glutamate receptor antagonists". Journal of Medicinal Chemistry. 47 (18): 4570–87. doi:10.1021/jm0400294. PMID 15317467.,
b) Yasuhara A, Nakamura M, Sakagami K, Shimazaki T, Yoshikawa R, Chaki S, et al. (June 2006). "Prodrugs of 3-(3,4-dichlorobenzyloxy)-2-amino-6-fluorobicyclo[3.1.0]hexane-2,6-dicarboxylic acid (MGS0039): a potent and orally active group II mGluR antagonist with antidepressant-like potential". Bioorganic & Medicinal Chemistry. 14 (12): 4193–207. doi:10.1016/j.bmc.2006.01.060. PMID 16487713.,
c) Yasuhara A, Sakagami K, Yoshikawa R, Chaki S, Nakamura M, Nakazato A (May 2006). "Synthesis, in vitro pharmacology, and structure-activity relationships of 2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylic acid derivatives as mGluR2 antagonists". Bioorganic & Medicinal Chemistry. 14 (10): 3405–20. doi:10.1016/j.bmc.2005.12.061. PMID 16431115. - ^ Woltering TJ, Adam G, Huguenin P, Wichmann J, Kolczewski S, Gatti S, et al. (February 2008). "Asymmetric synthesis and receptor pharmacology of the group II mGlu receptor ligand (1S,2R,3R,5R,6S)-2-amino-3-hydroxy-bicyclo[3.1.0]hexane-2,6-dicarboxylic acid-HYDIA". ChemMedChem. 3 (2): 323–35. doi:10.1002/cmdc.200700226. PMID 18058780. S2CID 1695024.
- ^ Wenthur CJ, Morrison R, Felts AS, Smith KA, Engers JL, Byers FW, et al. (June 2013). "Discovery of (R)-(2-fluoro-4-((-4-methoxyphenyl)ethynyl)phenyl) (3-hydroxypiperidin-1-yl)methanone (ML337), an mGlu3 selective and CNS penetrant negative allosteric modulator (NAM)". Journal of Medicinal Chemistry. 56 (12): 5208–12. doi:10.1021/jm400439t. PMC 3769689. PMID 23718281.
- ^ Hemstapat K, Da Costa H, Nong Y, Brady AE, Luo Q, Niswender CM, et al. (July 2007). "A novel family of potent negative allosteric modulators of group II metabotropic glutamate receptors". The Journal of Pharmacology and Experimental Therapeutics. 322 (1): 254–64. doi:10.1124/jpet.106.117093. PMID 17416742. S2CID 3820477.
- ^ Engers JL, Bollinger KA, Weiner RL, Rodriguez AL, Long MF, Breiner MM, et al. (September 2017). "Design and Synthesis of N-Aryl Phenoxyethoxy Pyridinones as Highly Selective and CNS Penetrant mGlu3 NAMs". ACS Medicinal Chemistry Letters. 8 (9): 925–930. doi:10.1021/acsmedchemlett.7b00249. PMC 5601378. PMID 28947938.
- ^ Woltering TJ, Wichmann J, Goetschi E, Adam G, Kew JN, Knoflach F, et al. (April 2008). "Synthesis and characterization of 1,3-dihydro-benzo[b][1,4]diazepin-2-one derivatives: Part 3. New potent non-competitive metabotropic glutamate receptor 2/3 antagonists". Bioorganic & Medicinal Chemistry Letters. 18 (8): 2725–9. doi:10.1016/j.bmcl.2008.02.076. PMID 18374569.
- ^ Zinni M, Mairesse J, Pansiot J, Fazio F, Iacovelli L, Antenucci N, Orlando R, Nicoletti F, Vaiman D, Baud O (January 2021). "mGlu3 receptor regulates microglial cell reactivity in neonatal rats". Journal of Neuroinflammation. 18 (1): 13. doi:10.1186/s12974-020-02049-z. PMC 7789385. PMID 33407565.
- ^ a b Hirbec H, Perestenko O, Nishimune A, Meyer G, Nakanishi S, Henley JM, Dev KK (May 2002). "The PDZ proteins PICK1, GRIP, and syntenin bind multiple glutamate receptor subtypes. Analysis of PDZ binding motifs". The Journal of Biological Chemistry. 277 (18): 15221–4. doi:10.1074/jbc.C200112200. hdl:2262/89271. PMID 11891216.
- ^ Flajolet M, Rakhilin S, Wang H, Starkova N, Nuangchamnong N, Nairn AC, Greengard P (December 2003). "Protein phosphatase 2C binds selectively to and dephosphorylates metabotropic glutamate receptor 3". Proceedings of the National Academy of Sciences of the United States of America. 100 (26): 16006–11. Bibcode:2003PNAS..10016006F. doi:10.1073/pnas.2136600100. PMC 307683. PMID 14663150.
Further reading[edit]
- Makoff A, Volpe F, Lelchuk R, Harrington K, Emson P (August 1996). "Molecular characterization and localization of human metabotropic glutamate receptor type 3". Brain Research. Molecular Brain Research. 40 (1): 55–63. doi:10.1016/0169-328X(96)00037-X. PMID 8840013.
- Emile L, Mercken L, Apiou F, Pradier L, Bock MD, Menager J, et al. (May 1996). "Molecular cloning, functional expression, pharmacological characterization and chromosomal localization of the human metabotropic glutamate receptor type 3". Neuropharmacology. 35 (5): 523–30. doi:10.1016/0028-3908(96)84622-3. PMID 8887960. S2CID 12697831.
- Corti C, Sala CF, Yang F, Corsi M, Xuereb JH, Ferraguti F (December 2000). "Genomic organization of the human metabotropic glutamate receptor subtype 3". Journal of Neurogenetics. 14 (4): 207–25, 271. doi:10.3109/01677060009084499. PMID 11342382. S2CID 44398182.
- Corti C, Xuereb JH, Corsi M, Ferraguti F (August 2001). "Identification and characterization of the promoter region of the GRM3 gene". Biochemical and Biophysical Research Communications. 286 (2): 381–7. doi:10.1006/bbrc.2001.5391. PMID 11500049.
- Tomiyama M, Kimura T, Maeda T, Tanaka H, Furusawa K, Kurahashi K, Matsunaga M (August 2001). "Expression of metabotropic glutamate receptor mRNAs in the human spinal cord: implications for selective vulnerability of spinal motor neurons in amyotrophic lateral sclerosis". Journal of the Neurological Sciences. 189 (1–2): 65–9. doi:10.1016/S0022-510X(01)00561-5. PMID 11535235. S2CID 34762564.
- Rosemond E, Peltekova V, Naples M, Thøgersen H, Hampson DR (March 2002). "Molecular determinants of high affinity binding to group III metabotropic glutamate receptors". The Journal of Biological Chemistry. 277 (9): 7333–40. doi:10.1074/jbc.M110476200. PMID 11744707.
- Martí SB, Cichon S, Propping P, Nöthen M (January 2002). "Metabotropic glutamate receptor 3 (GRM3) gene variation is not associated with schizophrenia or bipolar affective disorder in the German population". American Journal of Medical Genetics. 114 (1): 46–50. doi:10.1002/ajmg.1624. PMID 11840505.
- Kitano J, Kimura K, Yamazaki Y, Soda T, Shigemoto R, Nakajima Y, Nakanishi S (February 2002). "Tamalin, a PDZ domain-containing protein, links a protein complex formation of group 1 metabotropic glutamate receptors and the guanine nucleotide exchange factor cytohesins". The Journal of Neuroscience. 22 (4): 1280–9. doi:10.1523/JNEUROSCI.22-04-01280.2002. PMC 6757580. PMID 11850456.
- Hirbec H, Perestenko O, Nishimune A, Meyer G, Nakanishi S, Henley JM, Dev KK (May 2002). "The PDZ proteins PICK1, GRIP, and syntenin bind multiple glutamate receptor subtypes. Analysis of PDZ binding motifs". The Journal of Biological Chemistry. 277 (18): 15221–4. doi:10.1074/jbc.C200112200. hdl:2262/89271. PMID 11891216.
- Fujii Y, Shibata H, Kikuta R, Makino C, Tani A, Hirata N, et al. (June 2003). "Positive associations of polymorphisms in the metabotropic glutamate receptor type 3 gene (GRM3) with schizophrenia". Psychiatric Genetics. 13 (2): 71–6. doi:10.1097/01.ypg.0000056682.82896.b0. PMID 12782962. S2CID 22535773.
- Aronica E, Gorter JA, Ijlst-Keizers H, Rozemuller AJ, Yankaya B, Leenstra S, Troost D (May 2003). "Expression and functional role of mGluR3 and mGluR5 in human astrocytes and glioma cells: opposite regulation of glutamate transporter proteins". The European Journal of Neuroscience. 17 (10): 2106–18. doi:10.1046/j.1460-9568.2003.02657.x. PMID 12786977. S2CID 23408003.
- Flajolet M, Rakhilin S, Wang H, Starkova N, Nuangchamnong N, Nairn AC, Greengard P (December 2003). "Protein phosphatase 2C binds selectively to and dephosphorylates metabotropic glutamate receptor 3". Proceedings of the National Academy of Sciences of the United States of America. 100 (26): 16006–11. Bibcode:2003PNAS..10016006F. doi:10.1073/pnas.2136600100. PMC 307683. PMID 14663150.
- Yao Y, Koo JC, Wells JW, Hampson DR (June 2004). "Expression of a truncated secreted form of the mGluR3 subtype of metabotropic glutamate receptor". Biochemical and Biophysical Research Communications. 319 (2): 622–8. doi:10.1016/j.bbrc.2004.05.032. PMID 15178451.
- Tang FR, Chia SC, Chen PM, Gao H, Lee WL, Yeo TS, et al. (2004). "Metabotropic glutamate receptor 2/3 in the hippocampus of patients with mesial temporal lobe epilepsy, and of rats and mice after pilocarpine-induced status epilepticus". Epilepsy Research. 59 (2–3): 167–80. doi:10.1016/j.eplepsyres.2004.04.002. PMID 15246118. S2CID 6205182.
- Egan MF, Straub RE, Goldberg TE, Yakub I, Callicott JH, Hariri AR, et al. (August 2004). "Variation in GRM3 affects cognition, prefrontal glutamate, and risk for schizophrenia". Proceedings of the National Academy of Sciences of the United States of America. 101 (34): 12604–9. Bibcode:2004PNAS..10112604E. doi:10.1073/pnas.0405077101. PMC 515104. PMID 15310849.
External links[edit]
- "Metabotropic Glutamate Receptors: mGlu3". IUPHAR Database of Receptors and Ion Channels. International Union of Basic and Clinical Pharmacology. Archived from the original on 2014-08-08. Retrieved 2008-12-05.
This article incorporates text from the United States National Library of Medicine, which is in the public domain.







