| APOE | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | APOE, AD2, APO-E, LDLCQ5, LPG, apolipoprotein E, ApoE4 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 107741 MGI: 88057 HomoloGene: 30951 GeneCards: APOE | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Apolipoprotein E (Apo-E) is a protein involved in the metabolism of fats in the body of mammals. A subtype is implicated in Alzheimer's disease and cardiovascular diseases.[5] It is encoded in humans by the gene APOE.
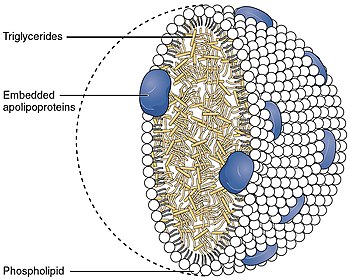
Apo-E belongs to a family of fat-binding proteins called apolipoproteins. In the circulation, it is present as part of several classes of lipoprotein particles, including chylomicron remnants, VLDL, IDL, and some HDL.[6] APOE interacts significantly with the low-density lipoprotein receptor (LDLR), which is essential for the normal processing (catabolism) of triglyceride-rich lipoproteins.[7] In peripheral tissues, APOE is primarily produced by the liver and macrophages, and mediates cholesterol metabolism. In the central nervous system, Apo-E is mainly produced by astrocytes and transports cholesterol to neurons[8] via APOE receptors, which are members of the low density lipoprotein receptor gene family.[9] Apo-E is the principal cholesterol carrier in the brain.[10] APOE qualifies as a checkpoint inhibitor of the classical complement pathway by complex formation with activated C1q.[11]
Evolution[edit]
Apolipoproteins are not unique to mammals. Many terrestrial and marine vertebrates have versions of them.[12] It is believed that APOE arose via gene duplications of APOC1 before the fish-tetrapod split c. 400 million years ago. Proteins similar in function have been found in choanoflagellates, suggesting that they are a very old class of proteins predating the dawn of all living animals.[13]
The three major human alleles (E4, E3, E2) arose after the primate-human split around 7.5 million years ago. These alleles are the by-product of non-synonymous mutations which led to changes in functionality. The first allele to emerge was E4. After the primate-human split, there were four amino acid changes in the human lineage, three of which had no effect on protein function (V174L, A18T, A135V). The fourth substitution (T61R) traded a threonine for an arginine altering the protein's functionality. This substitution occurred somewhere in the 6 million year gap between the primate-human split and the Denisovan-human split, since exactly the same substitutions were found in Denisovan APOE.[14]
About 220,000 years ago, a cysteine to arginine substitution took place at amino acid 112 (Cys112Arg) of the APOE4 gene, and this resulted in the E3 allele. Finally, 80,000 years ago, another arginine to cysteine substitution at amino acid 158 (Arg158Cys) of the APOE3 gene created the E2 allele.[15][13]
Structure[edit]
Gene[edit]
The gene, APOE, is mapped to chromosome 19 in a cluster with apolipoprotein C1 (APOC1) and the apolipoprotein C2 (APOC2). The APOE gene consists of four exons and three introns, totaling 3597 base pairs. APOE is transcriptionally activated by the liver X receptor (an important regulator of cholesterol, fatty acid, and glucose homeostasis) and peroxisome proliferator-activated receptor γ, nuclear receptors that form heterodimers with retinoid X receptors.[16] In melanocytic cells APOE gene expression may be regulated by MITF.[17]
Protein[edit]
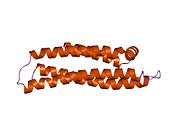
APOE is 299 amino acids long and contains multiple amphipathic α-helices. According to crystallography studies, a hinge region connects the N- and C-terminal regions of the protein. The N-terminal region (residues 1–167) forms an anti-parallel four-helix bundle such that the non-polar sides face inside the protein. Meanwhile, the C-terminal domain (residues 206–299) contains three α-helices which form a large exposed hydrophobic surface and interact with those in the N-terminal helix bundle domain through hydrogen bonds and salt-bridges. The C-terminal region also contains a low density lipoprotein receptor (LDLR)-binding site.[18]
Polymorphisms[edit]
| SNP: rs429358 | |
|---|---|
| Gene | ApoE |
| Chromosome | 19 |
| External databases | |
| Ensembl | Human SNPView |
| dbSNP | 429358 |
| HapMap | 429358 |
| SNPedia | 429358 |
APOE is polymorphic,[19][20] with three major alleles (epsilon 2, epsilon 3, and epsilon 4): APOE-ε2 (cys112, cys158), APOE-ε3 (cys112, arg158), and APOE-ε4 (arg112, arg158).[5][21][22] Although these allelic forms differ from each other by only one or two amino acids at positions 112 and 158,[23][24][25] these differences alter APOE structure and function.
There are several low-frequency polymorphisms of APOE. APOE5 comes in two subtypes E5f and E5s, based on migration rates. APOE5 E5f and APOE7 combined were found in 2.8% of Japanese males.[26][unreliable medical source] APOE7 is a mutation of APOE3 with two lysine residues replacing glutamic acid residues at positions 244 and 245.[27]
| Polymorphism | Worldwide allele frequency | Disease relevance |
|---|---|---|
| ε2 (rs7412-T, rs429358-T) | 8.4%[9] | This variant of the apoprotein binds poorly to cell surface receptors while E3 and E4 bind well.[28] E2 is associated with both increased and decreased risk for atherosclerosis. Individuals with an E2/E2 combination may clear dietary fat slowly and be at greater risk for early vascular disease and the genetic disorder type III hyperlipoproteinemia—94.4% of people with such disease are E2/E2 but only ~2% of E2/E2 develop it, so other environmental and genetic factors are likely to be involved (such as cholesterol in the diet and age).[29][30][31] E2 has also been implicated in Parkinson's disease,[32] but this finding was not replicated in a larger population association study.[33] |
| ε3 (rs7412-C, rs429358-T) | 77.9%[9] | This variant is considered the "neutral" APOE genotype. |
| ε4 (rs7412-C, rs429358-C) | 13.7%[9] |
E4 has been implicated in atherosclerosis,[34][35] Alzheimer's disease,[36][37] impaired cognitive function,[38][39] reduced hippocampal volume,[40] HIV,[41] faster disease progression in multiple sclerosis,[42][43] unfavorable outcome after traumatic brain injury,[44] ischemic cerebrovascular disease,[45] sleep apnea,[46][47] both the extension and shortening of telomeres,[48][49][50][51] reduced neurite outgrowth,[52] and COVID-19.[53] However, E4 has also been associated with enhanced vitamin D and calcium status,[54] higher fecundity,[55] protection against early childhood infection and malnutrition,[56] and decreased fetal, perinatal, and infant mortality.[57] |
Much remains to be learned about the APOE isoforms, including the interaction of other protective genes.[58] Indeed, the apolipoprotein ε4 isoform is more protective against cognitive decline than other isoforms in some cases,[58] so caution is advised before making determinant statements about the influence of APOE polymorphisms on cognition, development of Alzheimer's disease, cardiovascular disease, telomere shortening, etc. Many of the studies cited that purport these adverse outcomes are from single studies that have not been replicated and the research is based on unchecked assumptions about this isoform. As of 2007, there was no evidence that APOE polymorphisms influence cognition in younger age groups (other than possible increased episodic memory ability and neural efficiency in younger APOE4 age groups), nor that the APOE4 isoform places individuals at increased risk for any infectious disease.[59]
However, the association between the APOE4 allele and Alzheimer's disease has been shown to be weaker in minority groups differently compared to their Caucasian counterparts.[9] Hispanics/Latinos and African Americans who were homozygous for the APOE4 allele had 2.2 and 5.7 times the odds, respectively of developing Alzheimer's disease.[60][9] The APOE4 allele has an even stronger effect in East Asian populations, with Japanese populations have 33 times the odds compared to other populations.[61] Caucasians who were homozygous for the allele had 12.5 times the odds.[60][9]
Function[edit]
As a component of the lipoprotein lipid transport system, APOE facilitates the transport of lipids, fat-soluble vitamins, and cholesterol via the blood. It interacts with the LDL receptor to facilitate endocytosis of VLDL remnants. It is synthesized principally in the liver, but has also been found in other tissues such as the brain, kidneys, and spleen.[21] APOE synthesized in the liver associates with HDL which can then distribute it to newly formed VLDL or chylomicron particles to facilitate their eventual uptake by the liver.
In the nervous system, non-neuronal cell types, most notably astroglia and microglia, are the primary producers of APOE, while neurons preferentially express the receptors for APOE.[62] There are seven currently identified mammalian receptors for APOE which belong to the evolutionarily conserved LDLR family.[63]
APOE was initially recognized for its importance in lipoprotein metabolism and cardiovascular disease. Defects in APOE result in familial dysbetalipoproteinemia aka type III hyperlipoproteinemia (HLP III), in which increased plasma cholesterol and triglycerides are the consequence of impaired clearance of chylomicron, VLDL and LDL.[64][7] More recently, it has been studied for its role in several biological processes not directly related to lipoprotein transport, including Alzheimer's disease (AD), immunoregulation, and cognition.[5] Though the exact mechanisms remain to be elucidated, isoform 4 of APOE, encoded by an APOE allele, has been associated with increased calcium ion levels and apoptosis following mechanical injury.[65]
In the field of immune regulation, a growing number of studies point to APOE's interaction with many immunological processes, including suppressing T cell proliferation, macrophage functioning regulation, lipid antigen presentation facilitation (by CD1)[66] to natural killer T cell as well as modulation of inflammation and oxidation.[67] APOE is produced by macrophages and APOE secretion has been shown to be restricted to classical monocytes in PBMC, and the secretion of APOE by monocytes is down regulated by inflammatory cytokines and upregulated by TGF-beta.[68]
Clinical significance[edit]
Alzheimer's disease[edit]
As of 2012, the E4 variant was the largest known genetic risk factor for late-onset sporadic Alzheimer's disease (AD) in a variety of ethnic groups.[69] However, the E4 variant does not correlate with risk in every population. Nigerian people have the highest observed frequency of the APOE4 allele in world populations,[70] but AD is rare among them.[70][71] This may be due to their low cholesterol levels.[70][71][72][73] Caucasian and Japanese carriers of two E4 alleles have between 10 and 30 times the risk of developing AD by 75 years of age, as compared to those not carrying any E4 alleles. This may be caused by an interaction with amyloid.[74] Alzheimer's disease is characterized by build-ups of aggregates of the peptide beta-amyloid. Apolipoprotein E enhances proteolytic break-down of this peptide, both within and between cells. The isoform APOE-ε4 is not as effective as the others at promoting these reactions, resulting in increased vulnerability to AD in individuals with that gene variation.[75]
Recently, the amyloid hypothesis of Alzheimer's disease has been questioned, and an article in Science claimed that "Just as removing smoke does not extinguish a fire, reducing amyloid plaques may not affect the course of Alzheimer's disease."[76] The role that the E4 variant carries can still be fully explained even in the absence of a valid amyloid hypothesis given the fact that reelin signaling emerges to be one of the key processes involved in Alzheimer's disease[77] and the E4 variant is shown to interact with ApoER2, one of the neuronal reelin receptors, thereby obstructing reelin signaling.[77]
Although 40–65% of AD patients have at least one copy of the ε4 allele, APOE4 is not a determinant of the disease. At least one-third of patients with AD are APOE4 negative and some APOE4 homozygotes never develop the disease. Yet those with two ε4 alleles have up to 20 times the risk of developing AD.[78] There is also evidence that the APOE2 allele may serve a protective role in AD.[79] Thus, the genotype most at risk for Alzheimer's disease and at an earlier age is APOE4,4. Using genotype APOE3,3 as a benchmark (with the persons who have this genotype regarded as having a risk level of 1.0) and for white populations only, individuals with genotype APOE4,4 have an odds ratio of 14.9 of developing Alzheimer's disease. Individuals with the APOE3,4 genotype face an odds ratio of 3.2, and people with a copy of the 2 allele and the 4 allele (APOE2,4), have an odds ratio of 2.6. Persons with one copy each of the 2 allele and the 3 allele (APOE2,3) have an odds ratio of 0.6. Persons with two copies of the 2 allele (APOE2,2) also have an odds ratio of 0.6.[80]
| Estimated worldwide human allele frequencies of APOE in Caucasian population[80] | ||||
| Allele | ε2 | ε3 | ε4 | |
|---|---|---|---|---|
| General frequency | 8.4% | 77.9% | 13.7% | |
| AD frequency | 3.9% | 59.4% | 36.7% | |
While ApoE4 has been found to greatly increase the odds that an individual will develop Alzheimer's, a 2002 study concluded, that in persons with any combination of APOE alleles, high serum total cholesterol and high blood pressure in mid-life are independent risk factors which together can nearly triple the risk that the individual will later develop AD.[73] Projecting from their data, some researchers have suggested that lowering serum cholesterol levels may reduce a person's risk for Alzheimer's disease, even if they have two ApoE4 alleles, thus reducing the risk from nine or ten times the odds of getting AD down to just two times the odds.[73]
Women are more likely to develop AD than men across most ages and APOE genotypes. Premorbid women with the ε4 allele have significantly more neurological dysfunction than men.[81]
APOE-ε4 increases the risk not only for AD but also for dementia in pure alpha-synucleinopathies.[82] The influence of APOE-ε4 on hippocampal atrophy was suggested to be more predominant early in the course of AD at milder stages prior to more widespread neurodegeneration.[83]
Atherosclerosis[edit]
Knockout mice that lack the apolipoprotein-E gene (APOE−/−) develop extreme hypercholesterolemia when fed a high-fat diet.[84]
Malaria[edit]
APOE−/− knockout mice show marked attenuation of cerebral malaria and increased survival, as well as decreased sequestration of parasites and T cells within the brain, likely due to protection of the blood–brain barrier.[85] Human studies have shown that the APOE2 polymorphism correlates with earlier infection, and APOE3/4 polymorphisms increase likelihood of severe malaria.[86]
Lyme disease[edit]
Borrelia burgdorferi, the causative agent of Lyme disease, is a host-adapted pathogen that acquires environmental cholesterol to form glycolipids for use in cell membrane maintenance. In one experiment in 2015, mice engineered with apoE deficiency were infected with Borrelia spirochetes. The knockout mice suffered from an increased spirochete burden in joints, as well as inflamed ankles, when compared with wild-type mice. This study suggests that apoE deficiency (and potentially other hyperlipidemias) may be a risk factor in the pathogenicity of Lyme disease.
Interactions[edit]
Interactive pathway map[edit]
Click on genes, proteins and metabolites below to link to respective articles. [§ 1]
- ^ The interactive pathway map can be edited at WikiPathways: "Statin_Pathway_WP430".
References[edit]
- ^ a b c GRCh38: Ensembl release 89: ENSG00000130203 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000002985 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ a b c Stolerman IP, ed. (2010). Encyclopedia of Psychopharmacology (Online ed.). Berlin: Springer. ISBN 978-3540686989.
- ^ Mahley RW, Weisgraber KH, Huang Y (April 2009). "Apolipoprotein E: structure determines function, from atherosclerosis to Alzheimer's disease to AIDS". Journal of Lipid Research. 50 (Suppl): S183–S188. doi:10.1194/jlr.R800069-JLR200. PMC 2674716. PMID 19106071.
- ^ a b "Entrez Gene: APOE apolipoprotein E".
- ^ Wang H, Kulas JA, Wang C, Holtzman DM, Ferris HA, Hansen SB (August 2021). "Regulation of beta-amyloid production in neurons by astrocyte-derived cholesterol". Proceedings of the National Academy of Sciences of the United States of America. 118 (33): 2020.06.18.159632. Bibcode:2021PNAS..11802191W. bioRxiv 10.1101/2020.06.18.159632. doi:10.1073/pnas.2102191118. PMC 8379952. PMID 34385305. S2CID 220044671.
- ^ a b c d e f g Liu CC, Liu CC, Kanekiyo T, Xu H, Bu G (February 2013). "Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy". Nature Reviews. Neurology. 9 (2): 106–118. doi:10.1038/nrneurol.2012.263. PMC 3726719. PMID 23296339.
- ^ Puglielli L, Tanzi RE, Kovacs DM (April 2003). "Alzheimer's disease: the cholesterol connection". Nature Neuroscience. 6 (4): 345–351. doi:10.1038/nn0403-345. PMID 12658281. S2CID 5407666.
- ^ Yin C, Ackermann S, Ma Z, Mohanta SK, Zhang C, Li Y, et al. (March 2019). "ApoE attenuates unresolvable inflammation by complex formation with activated C1q". Nature Medicine. 25 (3): 496–506. doi:10.1038/s41591-018-0336-8. PMC 6420126. PMID 30692699.
- ^ Babin PJ, Thisse C, Durliat M, Andre M, Akimenko MA, Thisse B (August 1997). "Both apolipoprotein E and A-I genes are present in a nonmammalian vertebrate and are highly expressed during embryonic development". Proceedings of the National Academy of Sciences of the United States of America. 94 (16): 8622–8627. Bibcode:1997PNAS...94.8622B. doi:10.1073/pnas.94.16.8622. PMC 23048. PMID 9238027.
- ^ a b Huebbe P, Rimbach G (August 2017). "Evolution of human apolipoprotein E (APOE) isoforms: Gene structure, protein function and interaction with dietary factors". Ageing Research Reviews. 37: 146–161. doi:10.1016/j.arr.2017.06.002. PMID 28647612. S2CID 3758905.
- ^ McIntosh AM, Bennett C, Dickson D, Anestis SF, Watts DP, Webster TH, et al. (2012). "The apolipoprotein E (APOE) gene appears functionally monomorphic in chimpanzees (Pan troglodytes)". PLOS ONE. 7 (10): e47760. Bibcode:2012PLoSO...747760M. doi:10.1371/journal.pone.0047760. PMC 3480407. PMID 23112842.
- ^ Finch CE, Stanford CB (March 2004). "Meat-adaptive genes and the evolution of slower aging in humans". The Quarterly Review of Biology. 79 (1): 3–50. doi:10.1086/381662. PMID 15101252. S2CID 14225962.
- ^ Chawla A, Boisvert WA, Lee CH, Laffitte BA, Barak Y, Joseph SB, et al. (January 2001). "A PPAR gamma-LXR-ABCA1 pathway in macrophages is involved in cholesterol efflux and atherogenesis". Molecular Cell. 7 (1): 161–171. doi:10.1016/S1097-2765(01)00164-2. PMID 11172721.
- ^ Hoek KS, Schlegel NC, Eichhoff OM, Widmer DS, Praetorius C, Einarsson SO, et al. (December 2008). "Novel MITF targets identified using a two-step DNA microarray strategy". Pigment Cell & Melanoma Research. 21 (6): 665–676. doi:10.1111/j.1755-148X.2008.00505.x. PMID 19067971. S2CID 24698373.
- ^ Phillips MC (September 2014). "Apolipoprotein E isoforms and lipoprotein metabolism". IUBMB Life. 66 (9): 616–623. doi:10.1002/iub.1314. PMID 25328986. S2CID 6159310.
- ^ Singh PP, Singh M, Mastana SS (2006). "APOE distribution in world populations with new data from India and the UK". Annals of Human Biology. 33 (3): 279–308. doi:10.1080/03014460600594513. PMID 17092867. S2CID 34696595.
- ^ Eisenberg DT, Kuzawa CW, Hayes MG (September 2010). "Worldwide allele frequencies of the human apolipoprotein E gene: climate, local adaptations, and evolutionary history". American Journal of Physical Anthropology. 143 (1): 100–111. doi:10.1002/ajpa.21298. PMID 20734437.
- ^ a b Baars HF, van der Smagt JJ, Doevandans PA (2011). Clinical Cardiogenetics. London: Springer. ISBN 978-1849964715.
- ^ Ghebranious N, Ivacic L, Mallum J, Dokken C (October 2005). "Detection of ApoE E2, E3 and E4 alleles using MALDI-TOF mass spectrometry and the homogeneous mass-extend technology". Nucleic Acids Research. 33 (17): e149. doi:10.1093/nar/gni155. PMC 1243648. PMID 16204452.
- ^ Online Mendelian Inheritance in Man (OMIM): APOE3 isoform, hyperlipoproteinemia, type III, autosomal recessive - 107741#0015
- ^ Online Mendelian Inheritance in Man (OMIM): APOE3 isoform, APOE, CYS112 and ARG158 - 107741#0001
- ^ Zuo L, van Dyck CH, Luo X, Kranzler HR, Yang BZ, Gelernter J (April 2006). "Variation at APOE and STH loci and Alzheimer's disease". Behavioral and Brain Functions. 2 (1): 13. doi:10.1186/1744-9081-2-13. PMC 1526745. PMID 16603077.
- ^ Yamanouchi Y, Arinami T, Tsuchiya S, Miyazaki R, Takaki H, Takano T, Hamaguchi H (September 1994). "Apolipoprotein E5 and E7 in apparently healthy Japanese males: frequencies and relation to plasma lipid levels". The Japanese Journal of Human Genetics. 39 (3): 315–324. doi:10.1007/BF01874050. PMID 7841442. S2CID 20361600.
- ^ Dong J, Balestra ME, Newhouse YM, Weisgraber KH (November 2000). "Human apolipoprotein E7:lysine mutations in the carboxy-terminal domain are directly responsible for preferential binding to very low density lipoproteins". Journal of Lipid Research. 41 (11): 1783–1789. doi:10.1016/S0022-2275(20)31971-4. PMID 11060347.
- ^ Weisgraber KH, Innerarity TL, Mahley RW (March 1982). "Abnormal lipoprotein receptor-binding activity of the human E apoprotein due to cysteine-arginine interchange at a single site". The Journal of Biological Chemistry. 257 (5): 2518–2521. doi:10.1016/S0021-9258(18)34954-8. PMID 6277903.
- ^ Breslow JL, Zannis VI, SanGiacomo TR, Third JL, Tracy T, Glueck CJ (November 1982). "Studies of familial type III hyperlipoproteinemia using as a genetic marker the apoE phenotype E2/2". Journal of Lipid Research. 23 (8): 1224–1235. doi:10.1016/S0022-2275(20)38060-3. PMID 7175379.
- ^ Feussner G, Feussner V, Hoffmann MM, Lohrmann J, Wieland H, März W (1998). "Molecular basis of type III hyperlipoproteinemia in Germany". Human Mutation. 11 (6): 417–423. doi:10.1002/(SICI)1098-1004(1998)11:6<417::AID-HUMU1>3.0.CO;2-5. PMID 9603433. S2CID 39103399.
- ^ Civeira F, Pocoví M, Cenarro A, Casao E, Vilella E, Joven J, et al. (December 1996). "Apo E variants in patients with type III hyperlipoproteinemia". Atherosclerosis. 127 (2): 273–282. doi:10.1016/S0021-9150(96)05969-2. PMID 9125318.
- ^ Huang X, Chen PC, Poole C (June 2004). "APOE-[epsilon]2 allele associated with higher prevalence of sporadic Parkinson disease". Neurology. 62 (12): 2198–2202. doi:10.1212/01.wnl.0000130159.28215.6a. PMID 15210882. S2CID 10445412.
- ^ Federoff M, Jimenez-Rolando B, Nalls MA, Singleton AB (May 2012). "A large study reveals no association between APOE and Parkinson's disease". Neurobiology of Disease. 46 (2): 389–392. doi:10.1016/j.nbd.2012.02.002. PMC 3323723. PMID 22349451.
- ^ Phillips MC (September 2014). "Apolipoprotein E isoforms and lipoprotein metabolism". IUBMB Life. 66 (9): 616–623. doi:10.1002/iub.1314. PMID 25328986. S2CID 6159310.
- ^ Mahley RW, Rall SC (2000). "Apolipoprotein E: far more than a lipid transport protein". Annual Review of Genomics and Human Genetics. 1: 507–537. doi:10.1146/annurev.genom.1.1.507. PMID 11701639.
- ^ Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al. (August 1993). "Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families". Science. 261 (5123): 921–923. Bibcode:1993Sci...261..921C. doi:10.1126/science.8346443. PMID 8346443.
- ^ Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD (March 1993). "Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease". Proceedings of the National Academy of Sciences of the United States of America. 90 (5): 1977–1981. Bibcode:1993PNAS...90.1977S. doi:10.1073/pnas.90.5.1977. PMC 46003. PMID 8446617.
- ^ Deary IJ, Whiteman MC, Pattie A, Starr JM, Hayward C, Wright AF, et al. (August 2002). "Cognitive change and the APOE epsilon 4 allele". Nature. 418 (6901): 932. doi:10.1038/418932a. hdl:1842/702. PMID 12198535. S2CID 4418270.
- ^ Farlow MR, He Y, Tekin S, Xu J, Lane R, Charles HC (November 2004). "Impact of APOE in mild cognitive impairment". Neurology. 63 (10): 1898–1901. doi:10.1212/01.wnl.0000144279.21502.b7. PMID 15557508. S2CID 21131734.
- ^ Saeed U, Desmarais P, Masellis M (August 2021). "The APOE ε4 variant and hippocampal atrophy in Alzheimer's disease and Lewy body dementia: a systematic review of magnetic resonance imaging studies and therapeutic relevance". Expert Review of Neurotherapeutics. 21 (8): 851–870. doi:10.1080/14737175.2021.1956904. PMID 34311631. S2CID 236451232.
- ^ Burt TD, Agan BK, Marconi VC, He W, Kulkarni H, Mold JE, et al. (June 2008). "Apolipoprotein (apo) E4 enhances HIV-1 cell entry in vitro, and the APOE epsilon4/epsilon4 genotype accelerates HIV disease progression". Proceedings of the National Academy of Sciences of the United States of America. 105 (25): 8718–8723. doi:10.1073/pnas.0803526105. PMC 2438419. PMID 18562290.
- ^ Chapman J, Vinokurov S, Achiron A, Karussis DM, Mitosek-Szewczyk K, Birnbaum M, et al. (February 2001). "APOE genotype is a major predictor of long-term progression of disability in MS". Neurology. 56 (3): 312–316. doi:10.1212/wnl.56.3.312. PMID 11171894. S2CID 40761206.
- ^ Schmidt S, Barcellos LF, DeSombre K, Rimmler JB, Lincoln RR, Bucher P, et al. (March 2002). "Association of polymorphisms in the apolipoprotein E region with susceptibility to and progression of multiple sclerosis". American Journal of Human Genetics. 70 (3): 708–717. doi:10.1086/339269. PMC 384947. PMID 11836653.
- ^ Friedman G, Froom P, Sazbon L, Grinblatt I, Shochina M, Tsenter J, et al. (January 1999). "Apolipoprotein E-epsilon4 genotype predicts a poor outcome in survivors of traumatic brain injury". Neurology. 52 (2): 244–248. doi:10.1212/wnl.52.2.244. PMID 9932938. S2CID 131908791.
- ^ McCarron MO, Delong D, Alberts MJ (October 1999). "APOE genotype as a risk factor for ischemic cerebrovascular disease: a meta-analysis". Neurology. 53 (6): 1308–1311. doi:10.1212/wnl.53.6.1308. PMID 10522889. S2CID 23443430.
- ^ Kadotani H, Kadotani T, Young T, Peppard PE, Finn L, Colrain IM, et al. (June 2001). "Association between apolipoprotein E epsilon4 and sleep-disordered breathing in adults". JAMA. 285 (22): 2888–2890. doi:10.1001/jama.285.22.2888. PMID 11401610.
- ^ Gottlieb DJ, DeStefano AL, Foley DJ, Mignot E, Redline S, Givelber RJ, Young T (August 2004). "APOE epsilon4 is associated with obstructive sleep apnea/hypopnea: the Sleep Heart Health Study". Neurology. 63 (4): 664–668. doi:10.1212/01.wnl.0000134671.99649.32. PMID 15326239. S2CID 12280483.
- ^ Jacobs EG, Kroenke C, Lin J, Epel ES, Kenna HA, Blackburn EH, Rasgon NL (February 2013). "Accelerated cell aging in female APOE-ε4 carriers: implications for hormone therapy use". PLOS ONE. 8 (2): e54713. Bibcode:2013PLoSO...854713J. doi:10.1371/journal.pone.0054713. PMC 3572118. PMID 23418430.
- ^ Wikgren M, Karlsson T, Nilbrink T, Nordfjäll K, Hultdin J, Sleegers K, et al. (February 2012). "APOE ε4 is associated with longer telomeres, and longer telomeres among ε4 carriers predicts worse episodic memory". Neurobiology of Aging. 33 (2): 335–344. doi:10.1016/j.neurobiolaging.2010.03.004. PMID 20395015. S2CID 27820056.
- ^ Honig LS, Schupf N, Lee JH, Tang MX, Mayeux R (August 2006). "Shorter telomeres are associated with mortality in those with APOE epsilon4 and dementia". Annals of Neurology. 60 (2): 181–187. doi:10.1002/ana.20894. PMID 16807921. S2CID 73120632.
- ^ Dhillon VS, Deo P, Chua A, Thomas P, Fenech M (September 2020). "Shorter Telomere Length in Carriers of APOE-ε4 and High Plasma Concentration of Glucose, Glyoxal and Other Advanced Glycation End Products (AGEs)". The Journals of Gerontology. Series A, Biological Sciences and Medical Sciences. 75 (10): 1894–1898. doi:10.1093/gerona/glz203. PMID 31541246.
- ^ Raber J (May 2008). "AR, apoE, and cognitive function". Hormones and Behavior. 53 (5): 706–715. doi:10.1016/j.yhbeh.2008.02.012. PMC 2409114. PMID 18395206.
- ^ Kuo CL, Pilling LC, Atkins JL, Masoli JA, Delgado J, Kuchel GA, Melzer D (October 2020). "APOE e4 Genotype Predicts Severe COVID-19 in the UK Biobank Community Cohort". The Journals of Gerontology. Series A, Biological Sciences and Medical Sciences. 75 (11): 2231–2232. doi:10.1093/gerona/glaa131. PMC 7314139. PMID 32451547.
- ^ Huebbe P, Nebel A, Siegert S, Moehring J, Boesch-Saadatmandi C, Most E, et al. (September 2011). "APOE ε4 is associated with higher vitamin D levels in targeted replacement mice and humans". FASEB Journal. 25 (9): 3262–3270. doi:10.1096/fj.11-180935. PMID 21659554. S2CID 22483645.
- ^ Jasienska G, Ellison PT, Galbarczyk A, Jasienski M, Kalemba-Drozdz M, Kapiszewska M, et al. (March 2015). "Apolipoprotein E (ApoE) polymorphism is related to differences in potential fertility in women: a case of antagonistic pleiotropy?". Proceedings. Biological Sciences. 282 (1803): 20142395. doi:10.1098/rspb.2014.2395. PMC 4345437. PMID 25673673.
- ^ Oriá RB, Patrick PD, Oriá MO, Lorntz B, Thompson MR, Azevedo OG, et al. (March 2010). "ApoE polymorphisms and diarrheal outcomes in Brazilian shanty town children". Brazilian Journal of Medical and Biological Research = Revista Brasileira de Pesquisas Medicas e Biologicas. 43 (3): 249–256. doi:10.1590/s0100-879x2010007500003. PMC 3057459. PMID 20401432.
- ^ Becher JC, Keeling JW, Bell J, Wyatt B, McIntosh N (August 2008). "Apolipoprotein E e4 and its prevalence in early childhood death due to sudden infant death syndrome or to recognised causes". Early Human Development. 84 (8): 549–554. doi:10.1016/j.earlhumdev.2008.01.002. PMID 18280677.
- ^ a b Sundermann EE, Wang C, Katz M, et al. Cholesteryl ester transfer protein genotype modifies the effect of apolipoprotein ε4 on memory decline in older adults. Neurobiol Aging. 2016;41:200.e7-200.e12. doi:10.1016/j.neurobiolaging.2016.02.006
- ^ Mondadori CR, de Quervain DJ, Buchmann A, Mustovic H, Wollmer MA, Schmidt CF, et al. (August 2007). "Better memory and neural efficiency in young apolipoprotein E epsilon4 carriers". Cerebral Cortex. 17 (8): 1934–1947. doi:10.1093/cercor/bhl103. hdl:20.500.11850/5720. PMID 17077159.
- ^ a b Huynh RA, Mohan C (2017). "Alzheimer's Disease: Biomarkers in the Genome, Blood, and Cerebrospinal Fluid". Frontiers in Neurology. 8: 102. doi:10.3389/fneur.2017.00102. PMC 5357660. PMID 28373857.
- ^ Miyashita A, Kikuchi M, Hara N, Ikeuchi T (March 2023). "Genetics of Alzheimer's disease: an East Asian perspective". Journal of Human Genetics. 68 (3): 115–124. doi:10.1038/s10038-022-01050-z. ISSN 1435-232X. PMC 9968656. PMID 35641666.
- ^ Zhang Z, Mu J, Li J, Li W, Song J (January 2013). "Aberrant apolipoprotein E expression and cognitive dysfunction in patients with poststroke depression". Genetic Testing and Molecular Biomarkers. 17 (1): 47–51. doi:10.1089/gtmb.2012.0253. PMC 3525887. PMID 23171142.
- ^ Rogers JT, Weeber EJ (August 2008). "Reelin and apoE actions on signal transduction, synaptic function and memory formation". Neuron Glia Biology. 4 (3): 259–270. doi:10.1017/S1740925X09990184. PMID 19674510.
- ^ Sacks FM (February 2015). "The crucial roles of apolipoproteins E and C-III in apoB lipoprotein metabolism in normolipidemia and hypertriglyceridemia". Current Opinion in Lipidology. 26 (1): 56–63. doi:10.1097/MOL.0000000000000146. PMC 4371603. PMID 25551803.
- ^ Jiang L, Zhong J, Dou X, Cheng C, Huang Z, Sun X (August 2015). "Effects of ApoE on intracellular calcium levels and apoptosis of neurons after mechanical injury". Neuroscience. 301: 375–383. doi:10.1016/j.neuroscience.2015.06.005. PMID 26073697. S2CID 42716198.
- ^ van den Elzen P, Garg S, León L, Brigl M, Leadbetter EA, Gumperz JE, et al. (October 2005). "Apolipoprotein-mediated pathways of lipid antigen presentation". Nature. 437 (7060): 906–910. Bibcode:2005Natur.437..906E. doi:10.1038/nature04001. PMID 16208376. S2CID 3109596.
- ^ Zhang HL, Wu J, Zhu J (2010). "The role of apolipoprotein E in Guillain-Barré syndrome and experimental autoimmune neuritis". Journal of Biomedicine & Biotechnology. 2010: 357412. doi:10.1155/2010/357412. PMC 2825561. PMID 20182542.
- ^ Braesch-Andersen S, Paulie S, Smedman C, Mia S, Kumagai-Braesch M (2013). "ApoE production in human monocytes and its regulation by inflammatory cytokines". PLOS ONE. 8 (11): e79908. Bibcode:2013PLoSO...879908B. doi:10.1371/journal.pone.0079908. PMC 3828220. PMID 24244577.
- ^ Sadigh-Eteghad S, Talebi M, Farhoudi M (October 2012). "Association of apolipoprotein E epsilon 4 allele with sporadic late onset Alzheimer's disease. A meta-analysis". Neurosciences. 17 (4): 321–326. PMID 23022896.
- ^ a b c Sepehrnia B, Kamboh MI, Adams-Campbell LL, Bunker CH, Nwankwo M, Majumder PP, Ferrell RE (October 1989). "Genetic studies of human apolipoproteins. X. The effect of the apolipoprotein E polymorphism on quantitative levels of lipoproteins in Nigerian blacks". American Journal of Human Genetics. 45 (4): 586–591. PMC 1683508. PMID 2491016.
- ^ a b Notkola IL, Sulkava R, Pekkanen J, Erkinjuntti T, Ehnholm C, Kivinen P, et al. (1998-01-01). "Serum total cholesterol, apolipoprotein E epsilon 4 allele, and Alzheimer's disease". Neuroepidemiology. 17 (1): 14–20. doi:10.1159/000026149. PMID 9549720. S2CID 71543885.
- ^ Petanceska SS, DeRosa S, Sharma A, Diaz N, Duff K, Tint SG, et al. (2003-01-01). "Changes in apolipoprotein E expression in response to dietary and pharmacological modulation of cholesterol". Journal of Molecular Neuroscience. 20 (3): 395–406. doi:10.1385/JMN:20:3:395. PMID 14501024. S2CID 35969696.
- ^ a b c Kivipelto M, Helkala EL, Laakso MP, Hänninen T, Hallikainen M, Alhainen K, et al. (August 2002). "Apolipoprotein E epsilon4 allele, elevated midlife total cholesterol level, and high midlife systolic blood pressure are independent risk factors for late-life Alzheimer disease". Annals of Internal Medicine. 137 (3): 149–155. doi:10.7326/0003-4819-137-3-200208060-00006. PMID 12160362. S2CID 23780605.
- ^ Wisniewski T, Frangione B (February 1992). "Apolipoprotein E: a pathological chaperone protein in patients with cerebral and systemic amyloid". Neuroscience Letters. 135 (2): 235–238. doi:10.1016/0304-3940(92)90444-C. PMID 1625800. S2CID 8839627.
- ^ Jiang Q, Lee CY, Mandrekar S, Wilkinson B, Cramer P, Zelcer N, et al. (June 2008). "ApoE promotes the proteolytic degradation of Abeta". Neuron. 58 (5): 681–693. doi:10.1016/j.neuron.2008.04.010. PMC 2493297. PMID 18549781.
- Lay summary in: "Mechanism Explains Link Between Apolipoprotein E And Alzheimer's Disease". ScienceDaily (Press release). June 13, 2008.
- ^ Thambisetty M, Howard R, Glymour MM, Schneider LS (October 2021). "Alzheimer's drugs: Does reducing amyloid work?". Science. 374 (6567): 544–545. Bibcode:2021Sci...374..544T. doi:10.1126/science.abl8366. PMID 34709898. S2CID 240152869.
- ^ a b Kovács KA (December 2021). "Relevance of a Novel Circuit-Level Model of Episodic Memories to Alzheimer's Disease". International Journal of Molecular Sciences. 23 (1): 462. doi:10.3390/ijms23010462. PMC 8745479. PMID 35008886.
- ^ Hauser PS, Ryan RO (October 2013). "Impact of apolipoprotein E on Alzheimer's disease". Current Alzheimer Research. 10 (8): 809–817. doi:10.2174/15672050113109990156. PMC 3995977. PMID 23919769.
- ^ Corder EH, Saunders AM, Risch NJ, Strittmatter WJ, Schmechel DE, Gaskell PC, et al. (June 1994). "Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease". Nature Genetics. 7 (2): 180–184. doi:10.1038/ng0694-180. PMID 7920638. S2CID 11137478.
- ^ a b Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, et al. (1997). "Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium". JAMA. 278 (16): 1349–1356. doi:10.1001/jama.1997.03550160069041. PMID 9343467.
- ^ Damoiseaux JS, Seeley WW, Zhou J, Shirer WR, Coppola G, Karydas A, et al. (June 2012). "Gender modulates the APOE ε4 effect in healthy older adults: convergent evidence from functional brain connectivity and spinal fluid tau levels". The Journal of Neuroscience. 32 (24): 8254–8262. doi:10.1523/JNEUROSCI.0305-12.2012. PMC 3394933. PMID 22699906.
- ^ Tsuang D, Leverenz JB, Lopez OL, Hamilton RL, Bennett DA, Schneider JA, et al. (February 2013). "APOE ε4 increases risk for dementia in pure synucleinopathies". JAMA Neurology. 70 (2): 223–228. doi:10.1001/jamaneurol.2013.600. PMC 3580799. PMID 23407718.
- ^ Saeed U, Desmarais P, Masellis M (2021-08-03). "The APOE ε4 variant and hippocampal atrophy in Alzheimer's disease and Lewy body dementia: a systematic review of magnetic resonance imaging studies and therapeutic relevance". Expert Review of Neurotherapeutics. 21 (8): 851–870. doi:10.1080/14737175.2021.1956904. ISSN 1473-7175. PMID 34311631. S2CID 236451232.
- ^ McNeill E, Channon KM, Greaves DR (March 2010). "Inflammatory cell recruitment in cardiovascular disease: murine models and potential clinical applications". Clinical Science. 118 (11): 641–655. doi:10.1042/CS20090488. PMID 20210786.
- ^ Kassa FA, Van Den Ham K, Rainone A, Fournier S, Boilard E, Olivier M (September 2016). "Absence of apolipoprotein E protects mice from cerebral malaria". Scientific Reports. 6: 33615. Bibcode:2016NatSR...633615K. doi:10.1038/srep33615. PMC 5028887. PMID 27647324.
- ^ Wozniak MA, Riley EM, Itzhaki RF (March 2004). "Apolipoprotein E polymorphisms and risk of malaria". Journal of Medical Genetics. 41 (3): 145–146. doi:10.1136/jmg.2003.014613. PMC 1735716. PMID 14985370.
Further reading[edit]
- Ashford JW (2004). "APOE genotype effects on Alzheimer's disease onset and epidemiology". Journal of Molecular Neuroscience. 23 (3): 157–165. doi:10.1385/JMN:23:3:157. PMID 15181244. S2CID 14864342.
- Beffert U, Danik M, Krzywkowski P, Ramassamy C, Berrada F, Poirier J (July 1998). "The neurobiology of apolipoproteins and their receptors in the CNS and Alzheimer's disease". Brain Research. Brain Research Reviews. 27 (2): 119–142. doi:10.1016/S0165-0173(98)00008-3. PMID 9622609. S2CID 28731779.
- Bennet AM, Di Angelantonio E, Ye Z, Wensley F, Dahlin A, Ahlbom A, et al. (September 2007). "Association of apolipoprotein E genotypes with lipid levels and coronary risk". JAMA. 298 (11): 1300–1311. doi:10.1001/jama.298.11.1300. PMID 17878422.
- Bocksch L, Stephens T, Lucas A, Singh B (December 2001). "Apolipoprotein E: possible therapeutic target for atherosclerosis". Current Drug Targets - Cardiovascular & Hematological Disorders. 1 (2): 93–106. doi:10.2174/1568006013337944. PMID 12769659.
- de Knijff P, van den Maagdenberg AM, Frants RR, Havekes LM (1995). "Genetic heterogeneity of apolipoprotein E and its influence on plasma lipid and lipoprotein levels". Human Mutation. 4 (3): 178–194. doi:10.1002/humu.1380040303. PMID 7833947. S2CID 41959843.
- Gunzburg MJ, Perugini MA, Howlett GJ (December 2007). "Structural basis for the recognition and cross-linking of amyloid fibrils by human apolipoprotein E". The Journal of Biological Chemistry. 282 (49): 35831–35841. doi:10.1074/jbc.M706425200. PMID 17916554.
- Huang Y, Weisgraber KH, Mucke L, Mahley RW (2004). "Apolipoprotein E: diversity of cellular origins, structural and biophysical properties, and effects in Alzheimer's disease". Journal of Molecular Neuroscience. 23 (3): 189–204. doi:10.1385/JMN:23:3:189. PMID 15181247. S2CID 40135107.
- Itzhaki RF, Dobson CB, Shipley SJ, Wozniak MA (June 2004). "The role of viruses and of APOE in dementia". Annals of the New York Academy of Sciences. 1019 (1): 15–18. Bibcode:2004NYASA1019...15I. doi:10.1196/annals.1297.003. PMID 15246985. S2CID 28979273.
- Kolbe D, da Silva NA, Dose J, Torres GG, Caliebe A, Krause-Kyora B, Nebel A (May 2023). "Current allele distribution of the human longevity gene APOE in Europe can mainly be explained by ancient admixture". Aging Cell. 22 (5). Wiley: e13819. doi:10.1111/acel.13819. PMC 10186601. PMID 36951219.
- Kolovou GD, Anagnostopoulou KK (August 2007). "Apolipoprotein E polymorphism, age and coronary heart disease". Ageing Research Reviews. 6 (2): 94–108. doi:10.1016/j.arr.2006.11.001. PMID 17224309. S2CID 35607578.
- Lambert JC, Amouyel P (August 2007). "Genetic heterogeneity of Alzheimer's disease: complexity and advances". Psychoneuroendocrinology. 32 (Suppl 1): S62–S70. doi:10.1016/j.psyneuen.2007.05.015. PMID 17659844. S2CID 8114580.
- Liu CC, Liu CC, Kanekiyo T, Xu H, Bu G (February 2013). "Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy". Nature Reviews. Neurology. 9 (2): 106–118. doi:10.1038/nrneurol.2012.263. PMC 3726719. PMID 23296339.
- Mahley RW, Ji ZS (January 1999). "Remnant lipoprotein metabolism: key pathways involving cell-surface heparan sulfate proteoglycans and apolipoprotein E". Journal of Lipid Research. 40 (1): 1–16. doi:10.1016/S0022-2275(20)33334-4. PMID 9869645.
- Mahley RW, Rall SC (2002). "Apolipoprotein E: far more than a lipid transport protein". Annual Review of Genomics and Human Genetics. 1 (1): 507–537. doi:10.1146/annurev.genom.1.1.507. PMID 11701639.
- Mahley RW (April 1988). "Apolipoprotein E: cholesterol transport protein with expanding role in cell biology". Science. 240 (4852): 622–630. Bibcode:1988Sci...240..622M. doi:10.1126/science.3283935. PMID 3283935.
- Masterman T, Hillert J (June 2004). "The telltale scan: APOE epsilon4 in multiple sclerosis". The Lancet. Neurology. 3 (6): 331. doi:10.1016/S1474-4422(04)00763-X. PMID 15157846. S2CID 54404547.
- Moriyama K, Sasaki J, Matsunaga A, Arakawa F, Takada Y, Araki K, et al. (September 1992). "Apolipoprotein E1 Lys-146----Glu with type III hyperlipoproteinemia". Biochimica et Biophysica Acta. 1128 (1): 58–64. doi:10.1016/0005-2760(92)90257-V. PMID 1356443.
- Parasuraman R, Greenwood PM, Sunderland T (April 2002). "The apolipoprotein E gene, attention, and brain function". Neuropsychology. 16 (2): 254–274. doi:10.1037/0894-4105.16.2.254. PMC 1350934. PMID 11949718.
- Raber J (2007). "Role of apolipoprotein E in anxiety". Neural Plasticity. 2007: 91236. doi:10.1155/2007/91236. PMC 1940061. PMID 17710250.
- Roses AD, Einstein G, Gilbert J, Goedert M, Han SH, Huang D, et al. (January 1996). "Morphological, biochemical, and genetic support for an apolipoprotein E effect on microtubular metabolism". Annals of the New York Academy of Sciences. 777 (1): 146–157. Bibcode:1996NYASA.777..146R. doi:10.1111/j.1749-6632.1996.tb34413.x. PMID 8624078. S2CID 9145181.
- Strittmatter WJ, Roses AD (May 1995). "Apolipoprotein E and Alzheimer disease". Proceedings of the National Academy of Sciences of the United States of America. 92 (11): 4725–4727. Bibcode:1995PNAS...92.4725S. doi:10.1073/pnas.92.11.4725. PMC 41779. PMID 7761390.
- Utermann G, Pruin N, Steinmetz A (January 1979). "Polymorphism of apolipoprotein E. III. Effect of a single polymorphic gene locus on plasma lipid levels in man". Clinical Genetics. 15 (1): 63–72. doi:10.1111/j.1399-0004.1979.tb02028.x. PMID 759055. S2CID 34127430.
- Ye J (August 2007). "Reliance of host cholesterol metabolic pathways for the life cycle of hepatitis C virus". PLOS Pathogens. 3 (8): e108. doi:10.1371/journal.ppat.0030108. PMC 1959368. PMID 17784784.
External links[edit]
- Apolipoproteins+E at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- apoe4.info – website for APOE-epsilon-4 carriers
- Human APOE genome location and APOE gene details page in the UCSC Genome Browser.
- Overview of all the structural information available in the PDB for UniProt: P02649 (Apolipoprotein E) at the PDBe-KB.